Summary
The main goal of this Marie-Curie project was to improve our understanding of the human central nervous system (CNS) by extending our knowledge of the white-matter micro-scale composition, which is integral to brain function. Information flow in the brain is conveyed from the periphery via white matter pathways in the spinal cord to the cortex. However, it is currently not known to which degree the micro-scale composition of these pathways (e.g. myelin-sheath thickness or axonal diameter) determines the performance of functional networks associated with the cortex.
The goal of this proposal was to develop a novel computational in-vivo MRI method, which we call Mesoscopic White-Matter magnetic resonance Imaging (MWMI). MWMI will allow the assessment of 4 specific micro-scale metrics at high spatial resolution: myelin, water concentration, axonal density and the ratio between inner and outer fiber diameters (g-ratio) - a surrogate measure for the conductance speed in a fiber. Conventional quantitative MRI (qMRI), such as Diffusion Tensor Imaging (DTI), can detect microstructural changes but do not provide any information on the origin of these changes. In contrast, MWMI is capable of detecting microstructural changes and in addition provides insight into the underlying processes leading to these changes (e.g. whether pain leads to axonal reorganization or (de)myelination).
To main objectives of this project can be summarized as follows:
(i) To develop biophysical models that link the MR signal to the microscopic tissue composition.
(ii) To achieve high spatial resolution and integration of different MRI techniques.
(iii) To apply these MWMI methods to the pain circuit, which is a fundamental and well-described circuit
Main results
The biophysical models developed during this project describe the following qMRI techniques: (a) diffusion MRI, (b) quantitative multi-parameter-mapping (MPM) data.
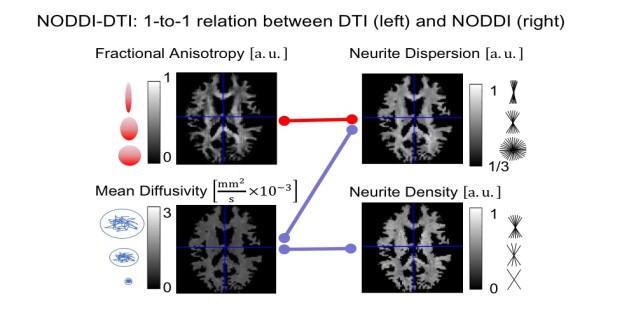
(a) We developed a new biophysical model, dubbed NODDI-DTI (see Figure), enabling the estimation of neurite density from commonly used diffusion tensor imaging (DTI) measurements (Edwards et al., 2017). NODDI-DTI allows to interpret DTI results in terms of the biophysical compartments of NODDI (e.g. are the observed changes in FA map due to changes in neurite density or fiber dispersion?). Moreover, we compared two different diffusion techniques for estimating the axonal compartment, namely double-diffusion encoding and standard Stejskal-Tanner diffusion encoding (Mohammadi et al., 2017b).
(b) We compared the comparability and repeatability of four different qMRI techniques to estimate the myelin (proton density and magnetization transfer imaging from MPMs) and axonal (NODDI and tract-fiber density from diffusion MRI) compartments for g-ratio weighted imaging (Ellerbrock and Mohammadi, 2018).
The image processing methods developed to improve data quality and thereby enabling integration of different qMRI techniques are:
(a) A new method for processing phase-reversed spin-echo EPI images in diffusion MRI, including Rician bias correction, motion and distortion correction, adaptive denoising, and efficient combination of blip-reversed spin-echo EPI data (Ellerbrock and Mohammadi, 2018).
(b) A novel method for detecting and removing corrupted DTI data in the spinal cord at the group level (David et al., 2017).
(c) Finally, we started working on a novel adaptive denoising method for quantitative multi-parameter-mapping (MPM) data that pools local data points with similar MR signal features and thus greatly preserves the effective spatial resolution (Tabelow et al., 2016; Mohammadi et al., 2017a).
To enable high spatial resolution for neuroscience (a) and clinical (b) usage, we developed efficient imaging protocols:
(a) The FLASH-based MPM sequence (Lutti et al., 2010, 2012; Weiskopf et al., 2013; Papp et al., 2016) was used for the acquisition of four high-resolution quantitative parameter maps (proton density, longitudinal and apparent transversal relaxation rate, and magnetization transfer maps) at 800 microns in only 25 minutes; the combination of parallel (Griswold et al., 2002) and multi-band (Moeller et al., 2010; Feinberg et al., 2010; Xu et al., 2013) imaging enabled the acquisition of multi-shell diffusion MRI data (b=0,1000,2000 s/mm2) at 1.6 mm isotropic resolution. Note that when acquiring the data we had access to a TIM TRIO MR system with a moderate gradient system (max. gradient amplitude 40mT/m). Using a new system (such as our new PRISMA-fit MR system, max. gradient amplitude 80mT/m), the diffusion MRI measurements could be done in a fraction of time or at a higher resolution.
(b) The time-efficient protocol for spinal cord DTI has been used in spinal cord injury studies (Huber et al., accepted; Grabher et al., 2016, 2017), and is now routinely used at the Spinal Cord Injury Center, University of Zürich.
During this project, we acquired behavioral and multi-modal qMRI data (incl. multi-shell diffusion MRI and MPMs) to investigate whether nociceptive long-term habituation will affect human brains microstructure. Although this project is still ongoing, first results have been obtained:
(a) First analyses of the behavioral data have shown that the healthy controls can be separated into strong and moderate sensitizers (Ellerbrock et al., 2016).
(b) First analyses of the MRI data have revealed the best processing and imaging protocol for assessing biophysical model parameters parameters such as the g-ratio (Ellerbrock and Mohammadi, 2018).
Finally, we are developing two open-source toolboxes that focused on processing and estimating MPM parameter maps (a) and processing DTI maps in the spinal cord (b):
(a) The hMRI toolbox (https://github.molgen.mpg.de/hMRI-group/Toolbox), which incorporates newest developments on modeling the MPM data, such as published in (Ellerbrock and Mohammadi, 2018). The corresponding toolbox abstract has been submitted to this years ISMRM conference in Paris.
(b) The ACID toolbox was extended by a spinal cord DTI branch. Currently, we are working on the toolbox paper. The toolbox will be released alongside with the paper, which is expected to be submitted in April, this year.
Impact and dissemination
The impact of the project from the scientific perspective can be summarized as follows:
- The NODDI-DTI method not only increases the understanding of how standard DTI metrics are related to biophysical properties (see Figure), but also will be one major pillar for achieving the goal of MWMI at mesoscopic spatial resolution. As compared to the original NODDI model, which requires a multi-shell diffusion MRI dataset (Zhang et al., 2012), NODDI-DTI enables the measurement of the neurite density and dispersion from standard single-shell DTI data. Therefore, it is currently one of the most SNR efficient biophysical models of the diffusion MRI signal to estimate neurite density.
- Our newly developed method for artifact correction in spinal cord DTI (David et al., 2017) reduces the artifact-induced variability across subjects and thus improves the statistical power for detecting group differences. In principle, this method can be extended to other model-based qMRI techniques, such as the MPM model.
This Marie-Curie project has inspired three new follow-up projects:
- To find the best biophysical model for estimating microstructure properties in the white matter, the grantee was awarded with an Emmy Noether Stipend Validating MRI-based in vivo histology by comparison with ex vivo histology (Funded by the DFG Emmy Noether Program ).
- To understand how biophysical models can be translated into clinical research, the grantee was awarded with a project as part of the hMRIofSCI ERA-NET Neuron consortium: Understanding the microstructural mechanisms of spinal cord injury (Funded by EU ERA-NET Neuron )
- To relate biophysical models to the human connectome, the grantee was awarded with a project as part of the Human Microstructural Connectomics projects:
In vivo and ex vivo characterization of the human microstructural connectome
(Funded by
DFG SPP 2041
).
This Marie-Curie project has inspired the following review articles and book chapters:
- A review about an integrative framework based on biophysical models that aims to characterize neurological disorders and minimize their impact on patients by considering functional interactions between supra-spinal, spinal and peripheral regions simultaneously (Freund et al., 2016).
- A review about computational neuroanatomy and microstructure imaging using magnetic resonance imaging (Mohammadi and Weiskopf, 2017).
- A book chapter about image analysis of quantitative MRI data in the second edition of the popular qMRI book Quantitative MRI of the Brain: Principles of Physical Measurement (Mohammadi and Callaghan, 2018)
The results of this Marie-Curie project have been disseminated through the following channels:
- Presentations at international conferences, including two oral presentations: one at the SFN 2016 (Ellerbrock and Mohammadi, 2016) and at the ISMRM 2017 (Ellerbrock and Mohammadi, 2017)
- The grantee has been an invited speaker on a presentation about computational MRI, biophysical models, and MRI-based in vivo histology. 39th Annual Meeting of the Japan Neuroscience Society 2016, Yokohama, Japan.
- At the ESMRMB lecture on Quantitative MRI for Characterising Brain Tissue Microstructure in Leipzig, 2016, the grantee has given a lecture about biophysical models using diffusion MRI.
- The methods developed in this Marie-Curie project are (or will become) available as open-source toolboxes: (a) the method for estimating a myelin biomarker (based on the PD map) is part of the hMRI toolbox and available here: https://github.molgen.mpg.de/hMRI-group/Toolbox (for details see (Ellerbrock and Mohammadi, 2018)), (b) the method for adaptive denoising MPM maps will become available after the paper has been published (for a preprint see (Mohammadi et al. 2017)), (c) the methods for artifact correction in spinal cord DTI will become available under www.diffusiontools.com , after the paper has been published (submission is planed around April).
References
Peer-reviewed journal papers / preprints
David G, Freund P, Mohammadi S (2017) The efficiency of retrospective artifact correction methods in improving the statistical power of between-group differences in spinal cord DTI . Neuroimage 158:296307.
Edwards LJ, Pine KJ, Ellerbrock I, Weiskopf N, Mohammadi S (2017) NODDI-DTI: Estimating Neurite Orientation and Dispersion Parameters from a Diffusion Tensor in Healthy White Matter . Front Neurosci 11:720.
Ellerbrock I, Mohammadi S (2018) Four in vivo g-ratio-weighted imaging methods: Comparability and repeatability at the group level . Hum Brain Mapp 39:2441.
Grabher P, Mohammadi S, Trachsler A, Friedl S, David G, Sutter R, Weiskopf N, Thompson AJ, Curt A, Freund P (2016) Voxel-based analysis of grey and white matter degeneration in cervical spondylotic myelopathy . Sci Rep 6:24636.
Grabher P, Mohammadi S, David G, Freund P (2017) Neurodegeneration in the Spinal Ventral Horn Prior to Motor Impairment in Cervical Spondylotic Myelopathy . J Neurotrauma 34:23292334.
Feinberg DA, Moeller S, Smith SM, Auerbach E, Ramanna S, Gunther M, Glasser MF, Miller KL, Ugurbil K, Yacoub E (2010) Multiplexed echo planar imaging for sub-second whole brain FMRI and fast diffusion imaging . PLoS ONE 5:e15710.
Freund P, Friston K, Thompson AJ, Stephan KE, Ashburner J, Bach DR, Nagy Z, Helms G, Draganski B, Mohammadi S, Schwab ME, Curt A, Weiskopf N (2016) Embodied neurology: an integrative framework for neurological disorders . Brain 139:18551861.
Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A (2002) Generalized autocalibrating partially parallel acquisitions (GRAPPA ) . Magn Reson Med 47:12021210.
Huber E, David G, Thompson A, Weiskopf N, Mohammadi S, Freund P (accepted) Dorsal and ventral horn atrophy is associated with clinical outcome after spinal cord injury. Neurology.
Lutti A, Hutton C, Finsterbusch J, Helms G, Weiskopf N (2010) Optimization and validation of methods for mapping of the radiofrequency transmit field at 3T . Magn Reson Med 64:229238.
Lutti A, Stadler J, Josephs O, Windischberger C, Speck O, Bernarding J, Hutton C, Weiskopf N (2012) Robust and fast whole brain mapping of the RF transmit field B1 at 7T . PLoS ONE 7:e32379.
Moeller S, Yacoub E, Olman CA, Auerbach E, Strupp J, Harel N, Ugurbil K (2010) Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI . Magn Reson Med 63:11441153.
Mohammadi S, Weiskopf N (2017) [ Computational neuroanatomy and microstructure imaging using magnetic resonance imaging ] . Nervenarzt 88:839849.
Mohammadi S, DAlonzo C, Ruthotto L, Polzehl J, Ellerbrock I, Callaghan MF, Weiskopf N, Tabelow K (2017a) Simultaneous adaptive smoothing of relaxometry and quantitative magnetization transfer mapping . Weierstrass Institute for Applied Analysis and Stochastics: Preprint 2432.
Mohammadi S, Callaghan M (2018) Quantitative MRI of the Brain: Principles of Physical Measurement , Second edition: M. Cercignani, N. G. Dowell, & P. S. Tofts, eds. Boca Raton, FL: CRC Press.
Papp D, Callaghan MF, Meyer H, Buckley C, Weiskopf N (2016) Correction of inter-scan motion artifacts in quantitative R1 mapping by accounting for receive coil sensitivity effects . Magn Reson Med 76:14781485.
Weiskopf N, Suckling J, Williams G, Correia MM, Inkster B, Tait R, Ooi C, Bullmore ET, Lutti A (2013) Quantitative multi-parameter mapping of R1, PD*, MT and R2* at 3T: a multi-center validation . Front Neurosci 7:95.
Xu J, Moeller S, Auerbach EJ, Strupp J, Smith SM, Feinberg DA, Yacoub E, Ugurbil K (2013) Evaluation of slice accelerations using multiband echo planar imaging at 3 T . Neuroimage 83:9911001.
Conference proceedings
Callaghan M, Pine K, Tabelow K, Polzeh J, Weiskopf N, Mohammadi S (2016) Mapping Higher Order Components of the GRE Signal Decay at 7T with Short TE Data through Adaptive Smoothing In Proc Intl Soc Magn Reson Med. 2016;24:1539.
Ellerbrock I, Mohammadi S (2016) Clinical usability of in vivo MR g-ratio mapping methods (San Diego, USA), 489.09.
Ellerbrock I, Mohammadi S, May A (2016) A model-based approach identifies strong and moderate sensitizers in a longitudinal pain paradigm In Proceedings of the 22nd Human Brain Mapping meeting, Geneva.
Ellerbrock I, Mohammadi S (2017) Comparing in vivo MR g-ratio mapping methods: accuracy and precision at the group level In Proc. Intl. Soc. Mag. Reson. Med. 25 (2017), abstract: 0284 .
Mohammadi S, Ellerbrock I, Edwards L (2017b) Biomarkers for fiber density: comparing Stejskal-Tanner diffusion encoding metrics with microscopic diffusion anisotropy from double-diffusion encoding imaging In Proc. Intl. Soc. Mag. Reson. Med. 25 (2017), abstract: 3382 .
Tabelow K, DAlonzo C, Polzehl J, Callaghan M, Ruthotto L, Weiskopf N, Mohammadi S (2016) How to achieve very high resolution quantitative MRI at 3T? In Proceedings of the 22nd Human Brain Mapping meeting, Geneva.