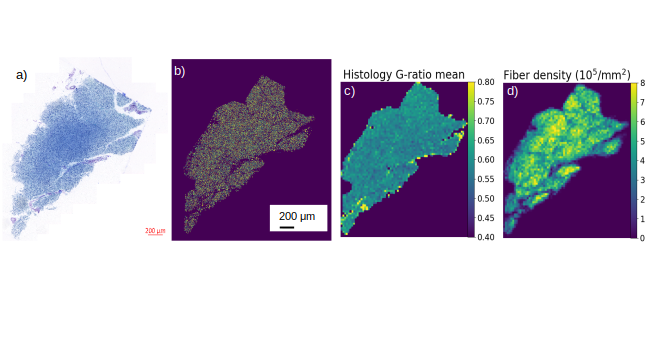
Figure 1: Microstructure map. a) Semi-thin section (1μm) from optic chiasm sample after stained with toluidine blue and scanned with an Axioscan Z1. b) Prediction mask of the entire slice. c, d) Examples for maps, representing averaged microstructure features of axons for the entire section: histological g-ratio (c) and fiber density (d).
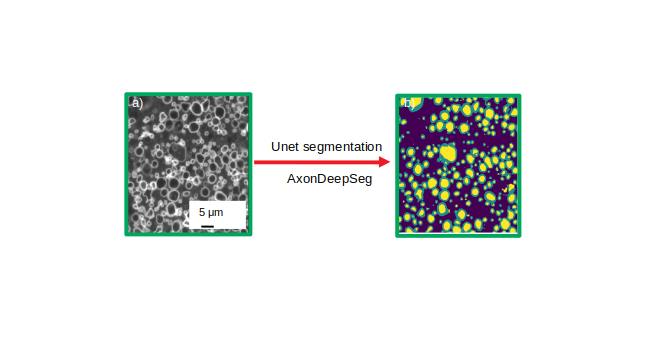
Figure 2: Unet segmentation using ADS . a) histology image (512x512 pixels), converted to grayscale. b) Prediction mask resulting from ADS, including 3 tissue classes: axon (yellow), myelin (green), and background (dark blue).
Deep learning for ex-vivo histology segmentation
Our group focus on developing biophysical models combined with quantitative MRI (aka in-vivo histology using MRI 1), to non-invasively assess the microstructure of brain tissue (e.g. axon diameters, axon and myelin densities, or the MR g-ratio, i.e. the relation between inner- and outer fiber diameter 2). This approach requires validation by comparison with a gold standard method, e.g. ex-vivo histology. Current validation attempts are often limited by one of the following challenges: (1) the ex-vivo histology method of choice is not quantitative and thus cannot be considered as a reliable gold standard, (2) the scale-gap between MRI, acquired at a macroscopic scale (~ millimeters), and ex-vivo histology, acquired at a microscopic scale (~ microns), hampers representative comparisons, (3) the tremendous amount of information from millions of axons need to be efficiently and reliably processed.
To obtain an accurate quantification (challenge 1), we take advantage of the submicroscopic resolution of light microscopy (with a point-spread function of ~250 nm) and an automatic segmentation of microscopic axonal features (diameters, myelin sheath thickness, densities). In a first attempt to overcome the scale-gap between ex-vivo histology and MRI (challenge 2), we are acquiring light microscopy of large tissue samples at macroscopic scales (~ few millimeters). To achieve an efficient and robust segmentation of a large number of ex-vivo histology images containing tremendous amount of information (challenge 3), we are using deep-learning-based methods.
This work aims at using and developing state-of-the-art segmentation techniques based on deep learning algorithm to generate macroscopic maps of microstructural features of axons such as density, diameter, or degree of myelination as assessed by the g-ratio see figure 1. With recent developments of deep learning, efficient methods for automatic segmentation of biological samples are available such as U-Net 3 and AxonDeepSeg 4, which uses convolution neural network to perform the segmentation task. We are developing a new pipelines using the neural network model based on a u-net architecture, adapting and improving (i.e. retraining of the network) its performance as compared to the original AxonDeepSeg implementation for efficient application to light microscopy images (figure 2, ISMRM 5). Additionally, we have implemented a novel post-processing sequence to access finer information at the fiber level such as fiber diameter and the g-ratio of individual axons, which were previously not available.
Acknowledgements: This project is part of the SPP 2041 (see also Microstructural Connectome ) and the Emmy Noether project (see also Validating MRI-based in-vivo histology ). The ex-vivo histology and light microscopy has been done by the group of Markus Morawski .
Reference:
1- Weiskopt N, Mohammadi S, Lutti A, Callaghan MF. Advances in MRI-based computational neuroanatomy: from morphometry to in-vivo histology (2015) Curr Opin Neurol 313-22
2- Mohammadi S, Carey D, Dick F, Diedrichsen J, Sereno MI, Reisert M, Callaghan MF, Weiskopf N (2015) Whole-Brain In-vivo Measurements of the Axonal G-Ratio in a Group of 37 Healthy Volunteers. Front Neurosci 9:441.
3- Ronnerberger O, Fischer P, Brox T. U-Net: Convolution Networks for Biomedical Image segementation (2015) MICCAI;234-241
4- Zaimi A, Wabartha M, Herman, V, Antonsanti P-L, Perone CS, Cohen-Adad J,. AxonDeepSeg: Automatic axon and myelin segmentation from microscopy data using convolutional neural network (2018) Sci. Rep. 8:3816
5- Tabarin T, Morozova M, Jaeger C, Rush H, Morawski M, Geyer S, Mohammadi S (accepted) Deep learning segmentation (AxonDeepSeg) to generate axonal-property map from ex vivo human optic chiasm using light microscopy In Proc. Intl. Soc. Mag. Reson. Med. 27 (2019), abstract: n.n.