Group Diagnostic
Research lab
N25
+49 (0) 40 7410 - 59827
Subversion, manipulation and exploitation of the host cell signalling machinery by pathogens is a common phenomenon during infection and the ability of pathogens to do so often largely defines their virulence and their ability to survive within the host. A well studied example of a pathogen, which affects host cell signalling at multiple levels in a sequential and highly complex way, is the gram-negative bacterium Yersinia, which includes the species Y. enterocolitica, Y. pseudotuberculosis and Y. pestis. Essential for virulence of all Yersiniae is an approximately 70 kb plasmid (pYV) carrying genes which encode components of a protein secretion system called Type III secretion system (TTSS) and six effectors YopE, YopT, YopO/YdkA, YopH, YopP and YopM. The TTSS is a highly complex needle-like multiprotein assembly, which enables Yersiniae to pierce through the plasma membrane of contacting host cells, most often cells of the innate immune system, and to inject the preformed Yops into the cytosol. The injected Yops immediately interfere heavily with several normal host cell signalling pathways to counteract infection, which ultimately prevents phagocytosis and cytokine production. This enables Yersiniae to remain extracellularly and replicate in lymphoid tissue.
This research group characterizes the intracellular activities of several Yops ofYersinia enterocolitica and their impact on innate immunity. Furthermore, novel interaction partners have been identified and are being characterized functionally and structurally.
Another focus of our research group is the identification of bacterial resistance against novel antibiotics with yet uncharacterized or poorly understood resistance mechanisms in clinical isolates. Resistance against multiple antibiotics simultaneously in gram-positive and gram-negative bacteria is a rapidly emerging problem. In recent years several new antibiotics have been brought on the market and serve as reserve antibiotics against these multiresistant strains. Unfortunately, as with all antibiotics so far resistance against these substances is emerging as they are used more frequently in the clinics. We collect clinical isolates, which exhibit unusual phenotypes for further analysis to understand the molecular basis, emergence and dissemination of resistance against diverse recently introduced antibiotics.
Project MALDI-TOF
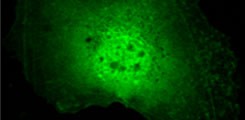
Characterization of YopM
Identification of novel interaction partners and analysis of manipulated signaling pathways.
Fig.1
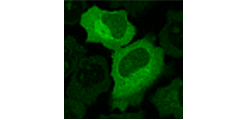
Antibiotic resistance
Characterization of bacterial resistance against novel or reserve antibiotics in clinical isolates.
Fig.2
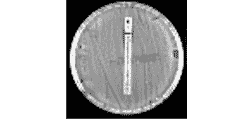
MALDI-TOF typing
Development and evaluation of mass spectrometry based typing strategies.
Fig.3
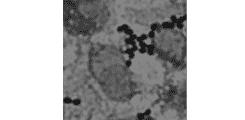
Rapid pathogen identification from blood culture fluid
Development and evaluation of methods for the preparation of microbial cells from blood culture fluid for MALDI-TOF identification.
Fig.4