Applied AI in Healthcare (AAI)
In the 'Applied AI in Healthcare' team, we strive to apply machine learning algorithms and "Deep Learning" models in clinical settings. Artificial intelligence, as we define it, encompasses methods that simulate human-like behavior for external observers.
In the specific sub-discipline of machine learning, our approach involves data analysis, constructing and adapting models that enable programs to "learn" through experience. We guide clinical projects benefiting from these methods from the early conceptual phase, providing advice on data collection and cleaning, to model development and critical evaluation. The foundation can be both routine medical diagnostics and study data, with a particular focus on classical tabular data and complex time series, such as sensor data or electronic health records.
In the Applied AI in Healthcare team, the generated algorithms and models are already seen as precursors to AI-based medical devices. Our focus remains on proximity to users in clinics, emphasizing the explainability of the models and their results (Explainable AI).
IAM-AI is a member of the
bAIome
.
Publications
Privacy Protection in Data Synthesis: Effects on Survival Analysis Performance
Beernink M, Gundler C
Stud Health Technol Inform. 2025;327:22-26.
Humane Endpoint Estimation in Laboratory Rodents Using Bayesian Time Series Forecasts
Blaß M, Grohmann C, Frenzel T, Gundler C
Stud Health Technol Inform. 2025;327:78-82.
Latent spaces of generative models for forensic age estimation: Evaluation of reliable support for forensic assessments
Chernysheva A, Gundler C, Wiederhold A, Jopp-van Well E, Heinemann A, Ondruschka B
RECHTSMEDIZIN. 2025;35(2):101–110.
Assessing the Generalizability of Foundation Models for the Recognition of Motor Examinations in Parkinson's Disease
Gundler C, Wiederhold A, Pötter-Nerger M
SENSORS-BASEL. 2025;25(17):5523.
Digitalizing Medical Forms Through Visual Question Answering: Are We There Yet?
Gundler C, Wiederhold A, Pötter-Nerger M
Stud Health Technol Inform. 2025;329:253-257.
Leveraging machine-learning techniques to detect recurrences in cancer registry data: A multi-registry validation study using German lung cancer data
Kusche H, Gundler C, Johanns O, Sauerberg M, Wicker T, Heinrichs V, Dettmer D, Goeken N, Langholz M, Luttmann S, Mazuch P, Pritzkuleit R, Rath N, Rausch K, Sha L, Sieber A, Katalinic A, Nennecke A
EUR J CANCER. 2025;227:115604.
Monitoring Hypomimia in Parkinson's Disease Using an E-Textile
Wiederhold A, Gundler C
Stud Health Technol Inform. 2025;327:999-1000.
Opportunities and Limitations of Wrist-Worn Devices for Dyskinesia Detection in Parkinson's Disease
Wiederhold A, Zhu Q, Spiegel S, Dadkhah A, Pötter-Nerger M, Langebrake C, Ückert F, Gundler C
SENSORS-BASEL. 2025;25(14):.
Unlocking the Potential of Secondary Data for Public Health Research: Retrospective Study With a Novel Clinical Platform
Gundler C, Gottfried K, Wiederhold A, Ataian M, Wurlitzer M, Gewehr J, Ückert F
INTERACT J MED RES. 2024;13:e51563.
Continuous, Learned Imputation of Missing Values in Parkinson's Disease
Gundler C, Pötter-Nerger M
Stud Health Technol Inform. 2024;316:654-658.
Improving Eye-Tracking Data Quality: A Framework for Reproducible Evaluation of Detection Algorithms
Gundler C, Temmen M, Gulberti A, Pötter-Nerger M, Ückert F
SENSORS-BASEL. 2024;24(9):2688.
Digitalizing Handwritten Digits of Patients with Parkinson's Disease Utilizing Consumer Hardware and Open-Source Software
Gundler C, Wiederhold A, Pötter-Nerger M
Stud Health Technol Inform. 2024;317:289-297.
Learning debiased graph representations from the OMOP common data model for synthetic data generation
Schulz N, Carus J, Wiederhold A, Johanns O, Peters F, Rath N, Rausch K, Holleczek , Katalinic A, Gundler C
BMC MED RES METHODOL. 2024;24(1):136.
An LLM-Based Visualization and Analysis Aid for a Secondary Use Clinical Data Platform
Spiegel S, Wendt T, Gundler C, Ückert F, Riemann L
Stud Health Technol Inform. 2024;316:1617-1621.
Mapping the Oncological Basis Dataset to the Standardized Vocabularies of a Common Data Model: A Feasibility Study
Carus J, Trübe L, Szczepanski P, Nürnberg S, Hees H, Bartels S, Nennecke A, Ückert F, Gundler C
CANCERS. 2023;15(16):.
Semi-Automated Mapping of German Study Data Concepts to an English Common Data Model
Chechulina A, Carus J, Breitfeld P, Gundler C, Hees H, Twerenbold R, Blankenberg S, Ückert F, Nürnberg S
APPL SCI-BASEL. 2023;13(14):8159.
A Unified Data Architecture for Assessing Motor Symptoms in Parkinson's Disease
Gundler C, Zhu Q, Trübe L, Dadkhah A, Gutowski T, Rosch M, Langebrake C, Nürnberg S, Baehr M, Ückert F
Stud Health Technol Inform. 2023;307:22-30.
Understanding Human-Computer Interactions in Restricted Clinical Environments
Kraus K, Trübe L, Gundler C
Stud Health Technol Inform. 2023;307:126-134.
Patient-individual 3D-printing of drugs within a machine-learning-assisted closed-loop medication management – Design and first results of a feasibility study
Langebrake C, Gottfried K, Dadkhah A, Eggert J, Gutowski T, Rosch M, Schönbeck N, Gundler C, Nürnberg S, Ückert F, Baehr M
Clinical eHealth. 2023;6:3-9.
Development of an immediate release excipient composition for 3D printing via direct powder extrusion in a hospital
Rosch M, Gutowski T, Baehr M, Eggert J, Gottfried K, Gundler C, Nürnberg S, Langebrake C, Dadkhah A
INT J PHARMACEUT. 2023;643:.
Introduction of MONOCLE – a software to reduce the workload and optimize the processes of the molecular tumor board at the University Hospital Hamburg-Eppendorf
Schmitz A, Lauk K, Heß K, Voß H, Fulla O, Schönbeck N, Ückert F, Gundler C, Riemann L
Ger Med Sci. 2023.
Letzte Aktualisierung aus dem FIS: 05.03.2026 - 04:37 Uhr
Current projects
-
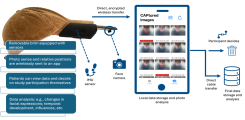
A Cap for Detecting Hypomimia
A Cap for Detecting Hypomimia
Parkinson's Disease (PD) is an incurable neurodegenerative disorder characterized by the loss of dopamine-producing cells in the substantia nigra. The resulting symptoms can significantly impact patients' quality of life. Current wearable technology is capable of identifying motor symptoms like tremor, but largely unexplored symptoms such as hypomimia, a reduction in facial expressiveness, remain difficult to assess. Present methods, such as in-clinic video recordings, do not allow for continuous monitoring in daily life, which is crucial for understanding disease progression and adjusting treatment strategies. Our team, in collaboration with the Movement Disorders and Deep Brain Stimulation Working Group led by PD Dr. med. Monika Pötter-Nerger, has developed and prototyped an innovative measurement instrument. This device can precisely monitor subtle facial muscle movements and wirelessly transmit the data to an iOS application. We are currently seeking dedicated medical doctoral candidates for the clinical management and initial exploratory analyses of this project. Interested individuals are encouraged to contact us via email. For further information, Christopher Gundler and Alexander Wiederhold are available as contacts.
-
Internet of Things and Parkinson's disease
Internet of Things and Parkinson's disease
To improve the quality of life for patients with Parkinson's disease, inpatient examinations and treatments are often conducted. During hospital stays, adjustments to the current treatment plan are prioritized. Accurate and personalized treatment is crucial for therapeutic success.
With the help of a specialized tablet application, patients have the opportunity to independently record relevant parameters of their current well-being and other non-motor symptoms. Similar to a digital diary, this allows for the collection of a wide range of disease-relevant data. This is complemented by the use of sensors that communicate with the tablet through locally networked systems, enabling the tracking of key vital parameters over time.
Through the use of these connected systems, we hope to gain new insights into the severity of Parkinson's disease symptoms. In a previous project, we have already collected comparable data as well as preliminary movement data and medication intake.