Cytometry and Cell Sorting Core Facility
The Cytometry and Cell Sorting Core Facility provides researchers with state-of-the-art technologies for flow cytometry and cell sorting. It is equipped with:
Analyzer (5)
-
Aurora (in HCTI room 04.2.022)
Aurora
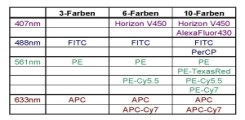
For high compexity applications there is the aurora as high end flowcytometer available in the core facility. It is equipped with the 3 standard lasers plus a yellowgreen and a uv-laser. Because this cytometer is a spectral cytometer (64 fluorescence channels + 3 scatter channels) you can use fluorochromes with similar fluorescence emissions.
The result of samples with high autofluorescence can be improved by autofluorescence extraction. -
A3 (in HCTI room 04.2.022)
The LSR Fortessa A3 is our highend flowcytometer for getting the most information out of your sample. The device provides the 3 standard lasers plus a yellow-green and an uv-laser. It is possible to use up to 29 parameters in your sample.
The device is equipped with the following lasers:
- 355 nm: UV laser
- 405 nm: Violet laser
- 488 nm: Blue Laser
- 561 nm: Green laser
- 640 nm: Red Laser
-
LSR Fortessa with 5 lasers (in CF1 room 00.068)
The LSR Fortessa 5L is for relief of the A3. Up to 18 fluorescences can be used in a sample.
The device is equipped with the following lasers:
- 350 nm: uv laser
- 405 nm: violet laser
- 488nm: blue laser
- 561 nm: green laser
- 640 nm: red laser
-
LSR Fortessa with 4 lasers (in CF1 room 00.068)
The LSR Fortessa allows, in addition to the blue, violet and red laser a yellow-green laser for better detection with the use of PE-dyes and red fluorescent proteins. Up to 17 fluorescences can be used in a sample.
The device is equipped with the following lasers:
- 405 nm: violet laser
- 488nm: blue Laser
- 561 nm: green laser
- 640 nm: red Laser
-
Quanteon (in CF1 room 0.068)
The quanteon is equipped with 4 lasers. The special feature of this device is the determination of absolute cell counts.
Furthermore the software provides analysis wizards for cell cycle and proliferation experiments.
The quanteon can be used independently for multicolour experiments or as a service.
The device is equipped with the following lasers:
- 405 nm: violet laser
- 488nm: blue Laser
- 561 nm: green laser
- 637 nm: red Laser
Sorter (3)
-
S6 SE (in HCTI room 04.2.22) 6-way sorting, spectral, 5 laser, untested primary human samples possible
The device is equipped with the following lasers:
- 355 nm: uv laser
- 405 nm: violet laser
- 488nm: blue laser
- 561 nm: green laser
- 637 nm: red laser
The S6 is equipped with the following new features:
- 6-way sorting
- spectral unmixing
-
AriaFusion (in CF1 room 00.068.1) 4-way sorting, 5 laser, untested primary human samples possible
The device is equipped with the following lasers:
- 355 nm: uv laser
- 405 nm: violet laser
- 488nm: blue laser
- 561 nm: green laser
- 640 nm: red laser
-
Aria IIIu (in CF1 room 0.068.1) 4-way sorting, 4 laser, untested primary human samples not possible
With this device one or more cell populations from the suspension can be sorted out (Note: not all cells can be sorted well, sometimes the populations are very sensitive). The sorted cells can be cultivated further.
The cell sorting on AriaIIIu is handled by employees of the Core Facility. The device is equipped with the following lasers:
- 407 nm: violet laser
- 488nm: Blue Laser
- 561 nm: green laser
- 633 nm: Red Laser
-
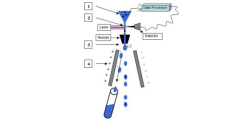
FACS Sorting - The principle
1. Cells are harvested by a sheath flow as beads strung on a string (hydrodynamic focusing)
2. In the laser intersection the analysis of the cells takes place.
3. The nozzle separates the jet into individual drops and charges the drops to be sorted on.
4. The charged drops are deflected in an electric field, and fall into the appropriate receptacle.
-
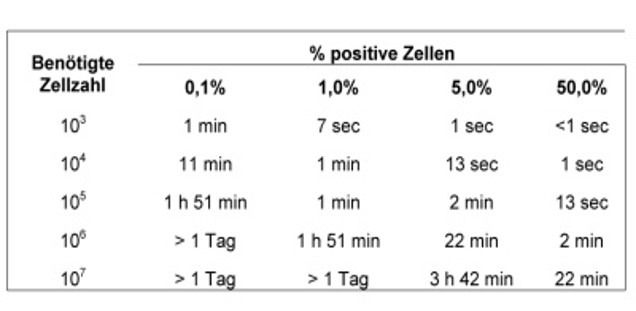
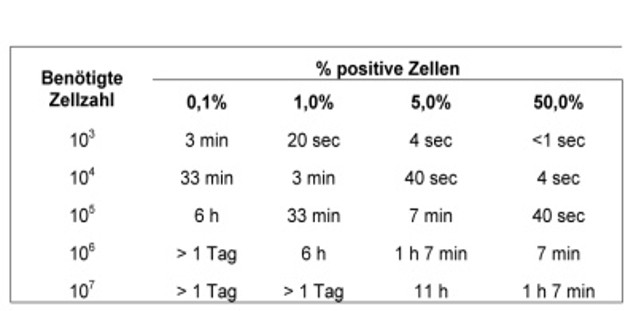
Sorting: duration
The following tables show how long a sorting operation. These are minimum hours, 20 minutes should be allocated for the setting of the device. The duration varies with the size of the cells.
-
Sorting: different sample collections
The following options are possible with the FACS Aria III and FACS Aria Fusion:
1) separated into 2 populations in 15 ml tubes
2) seperated into up to 4 populations in FACS-tubes or small reaction tubes
3) filing in microtiter plates (for example, 96-well)
Access, Security, and Booking
Before entering the laboratories, participation in the mandatory S2 safety training for our genetic engineering facilities is required. The training dates and registration are available and can be booked through our PPMS booking system.
All equipment reservations, safety trainings, user trainings, and billing are centrally managed via the PPMS system.
The analyzers may be used independently after successful completion of the training and confirmation of the terms of use.
Cell sorting is offered as a service.
Services
- Cell Sorting with the AriaIIIu, AriaFusion and S6 SE
- Sorting of untested human samples and bacteria (AriaFusion)
- Cell analysis with the A3, Quanteon, LSR Fortessa 4-laser and 5-laser and Aurora
- Determination of absolute cell counts on the Quanteon
- Analysis of cell cycle on the Quanteon
- High resolution images can be combined with quantifiable results (stained marker) on the imaging flow cytometer Image Stream
- Analysis of vesicles and microparticles on the Image Stream
- Provision of evaluation stations for independent use with the following software: DIVA, FLOWJo, NovoExpress, Infinicyt and Ideas Software
- Introductory training and support for new users on the analysis devices for independent use
- Training of users with many years of flow cytometry experience on the sorters, AriaIIIu and AriaFusion, for independent use
- Measurement of the latest fluorescent proteins possible through acquisition of new filters
- Regular training courses on flow cytometry in cooperation with the device manufacturers
- Scientific support: consultation on the development of a new panel and assistance with data analysis, e.g., using FlowJo (can be booked in PPMS under Tasking: Scientific Consultation).
Registration
The Cytometry and Cell Sorting Core Facility is open to all researchers at the UKE and other interested parties. Bookings can be made via the internet booking system provided. New users should contact the specified contact persons directly.
There is flat fee per hour of use, which is calculated on the basis of the required consumables. The usage fee for UKE employees is (as of september 2025):
• Aurora 35 € per hour
• A3 (5 laser) 25 € per hour
• LSR Fortessa (5 laser) 25 € per hour
• Quanteon (4 laser) 24 € per hour
• LSR Fortessa (4 laser) 22 € per hour
• AriaIIIu 60 € per hour
• AriaFusion 60 € per hour
• S6 SE 60 € per hour
The total usage time for each working group is determined on a quaterly basis and billed either via cost center or third-party funds. Please note the usage regulations for the individual devices.
Contact
Location CF1 (N27)
- lab: room 0.068 + 0.068.1 > tel. 52306 (best accessibility)
- office: room 0.066 > tel. 39677
Location HCTI (N25)
- lab: room 04.2.022 + 023.2 > tel. 50036
- office: room 04.2.037 > tel. 39644
facs@uke.de
_3_kontaktbild.jpg)
- Head of Science
- Cytometry and Cell Sorting Core Unit

- Medical-technical laboratory assistant
- Cytometry and Cell Sorting Core Unit

- Medical-technical laboratory assistant
- Cytometry and Cell Sorting Core Unit