In several research projects, the MVTV research team analyzes the molecular mechanisms of chronic viral infection, how these viruses establish persistence and contribute to pathogenesis including cancer. The research has a focus on pathogenesis mechanisms associated with the human polyomaviruses (hPyV), Merkel Cell Polyomavirus (MCPyV) and BKPyV and the development of intervention options, such as antivirals.
hPyVs are highly prevalent pathogens that establish lifelong asymptomatic persistence in the healthy immunocompetent host. These viruses are opportunistic pathogens and, under immunosuppression, contribute to uncontrolled replication and pathogenesis by reactivating persistence. MCPyV is the leading cause of Merkel Cell Carcinoma (MCC), a rare skin cancer, while PyV associated nephropathy (PVAN) is a consequence of BKV reactivation in kidney epithelial cells.
A major challenge in understanding these viruses and how to combat them is the restricted cell tropism and the associated limitation in infection and pathogenicity models.
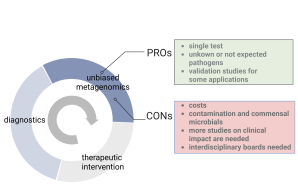
Another translational approach at the Institute is the use of high-throughput sequencing (incl. whole genome sequencing, targeted sequencing and metagenomics) for the detection of known pathogens and the identification of putative novel pathogens in diagnostic samples. In collaboration with the Leibniz Institute for Virology, LIV, Research Unit Virus Genomics and the Technology Unit Next-Generation Sequencing at the LIV, we develop and apply pre-analytical and bioinformatic methods for the detection, characterization and identification of pathogens in complex sequencing data.
Our research is conducted in our laboratories at the Campus Forschung II (N25).
Publications | chronic viral infection
Ohnezeit D, Huang J, Westerkamp U, Brinschwitz V, Schmidt C, Günther T, Czech-Sioli M, Weißelberg S, Schlemeyer T, Nakel J, Mai J, Schreiner S, Schneider C, Friedel CC, Schwanke H, Brinkmann MM, Grundhoff A, Fischer N (2024). Merkel cell polyomavirus small tumor antigen contributes to immune evasion by interfering with type I interferon signaling. PLoS Pathog. Aug 7;20(8):e1012426. doi: 10.1371/journal.ppat.1012426. Ohnezeit et al.
Albertini S, Martuscelli L, Borgogna C, Virdi S, Indenbirken D, Lo Cigno I, Griffante G, Calati F, Boldorini R, Fischer N#, Gariglio M# (2022) Cancer-Associated Fibroblasts Exert Proangiogenic Activity in Merkel Cell Carcinoma. J Invest Dermatol doi: 10.1016/j.jid.2022.12.006 (# corresponding authors). Albertini et al.
Schlemeyer T, Ohnezeit D, Virdi S, Korner C, Weisselberg S, Starzonek S, Schumacher U, Grundhoff A, Indenbirken D, Albertini, Fischer N (2022) Merkel Cell Carcinoma and Immune Evasion: Merkel Cell Polyomavirus Small T-Antigen‒Induced Surface Changes Can Be Reverted by Therapeutic Intervention. J Invest Dermatol 142: 3071-3081 e13 doi: 10.1016/j.jid.2022.04.029. Schlemeyer et al.
Czech-Sioli M, Gunther T, Therre M, Spohn M, Indenbirken D, Theiss J, Riethdorf S, Qi M, Alawi M, Wulbeck C, Fernandez-Cuesta I, Esmek F, Becker JC, Grundhoff A#, Fischer N# (2020) High-resolution analysis of Merkel Cell Polyomavirus in Merkel Cell Carcinoma reveals distinct integration patterns and suggests NHEJ and MMBIR as underlying mechanisms. PLoS Pathog 16: e1008562 doi: 10.1371/journal.ppat.1008562 (# corresponding author). Czech-Sioli et al.
Theiss JM, Gunther T, Alawi M, Neumann F, Tessmer U, Fischer N#, Grundhoff A# (2015) A Comprehensive Analysis of Replicating Merkel Cell Polyomavirus Genomes Delineates the Viral Transcription Program and Suggests a Role for mcv-miR-M1 in Episomal Persistence. PLoS Pathog 11: e1004974 doi: 10.1371/journal.ppat.1004974. Theiss et al.
Publications | metagenomics
Olearo F, Christner M, Lütgehetmann M, Aepfelbacher M, Fischer N, Rohde H. (2025) Revisiting diagnostics: microbial cell-free DNA-sequencing: addressing unmet challenges in implant-related cardiovascular infections. Clin Microbiol Infect. 31(4):510-512. doi: 10.1016/j.cmi.2024.11.019. Epub 2024 Nov 15. Olearo et al.
Czech-Sioli M, Gunther T, Robitaille A, Roggenkamp H, Buttner H, Indenbirken D, Christner M, Lutgehetmann M, Knobloch J, Aepfelbacher M, Grundhoff, Fischer N (2023) Integration of Sequencing and Epidemiologic Data for Surveillance of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infections in a Tertiary-Care Hospital. Clin Infect Dis 76: e263-e273 doi:10.1093/cid/ciac484. Czech-Sioli et al.
Baier C, Huang J, Reumann K, Indenbirken D, Thol F, Koenecke C, Ebadi E, Heim A, Bange FC, Haid S, Pietschmann T, Fischer N (2022) Target capture sequencing reveals a monoclonal outbreak of respiratory syncytial virus B infections among adult hematologic patients. Antimicrob Resist Infect Control 11: 88 doi: 10.1186/s13756-022-01120-z. Baier et al.
Heyer A, Gunther T, Robitaille A, Lutgehetmann M, Addo MM, Jarczak D, Kluge S, Aepfelbacher M, Schulze Zur Wiesch J, Fischer N# & Grundhoff A# (2022) Remdesivir-induced emergence of SARS-CoV2 variants in patients with prolonged infection. Cell Rep Med. 3 (9):100735 doi: 10.1016/j.xcrm.2022.100735 (# corresponding authors) Heyer et al.
Kemming J, Gundlach S, Panning M, Huzly D, Huang J, Lutgehetmann M, Pischke S, Schulze Zur Wiesch J, Emmerich F, Llewellyn-Lacey S, Price DA, Tanriver Y, Warnatz K, Boettler T, Thimme R, Hofmann M, Fischer N & Neumann-Haefelin C (2022) Mechanisms of CD8+ T-cell failure in chronic hepatitis E virus infection. J Hepatol 77: 978-990 doi: 10.1016/j.jhep.2022.05.019. Kemming et al.
Günther T, Czech-Sioli M, Indenbirken D, Robitaille A, Tenhaken P, Exner M, Ottinger M, Fischer N#, Grundhoff A#, Brinkmann MM#. SARS-CoV-2 outbreak investigation in a German meat processing plant. EMBO Mol Med. 2020 Dec 7;12(12):e13296. doi: 10.15252/emmm.202013296 (# corresponding authors). Günther et al.
Alawi M, Burkhardt L, Indenbirken D, Reumann K, Christopeit M, Kröger N, Lütgehetmann M, Aepfelbacher M, Fischer N, Grundhoff A. DAMIAN: an open source bioinformatics tool for fast, systematic and cohort based analysis of microorganisms in diagnostic samples. Sci Rep. 2019 Nov 14;9(1):16841. doi: 10.1038/s41598-019-52881-4.
Alawi et al.
Infection and Persistence
Gene expression changes during Merkel Cell Polyomavirus productive and persistent infection in skin organoids
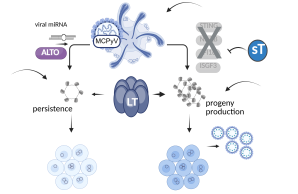
Due to the restricted cell tropism and the associated limitation in infection and pathogenicity models, the cell types of persistence and the mechanisms contributing to persistence for MCPyV remain to be elucidated. The primary objectives of this research project are to elucidate the molecular mechanisms that regulate productive Merkel cell polyomavirus (MCPyV) infection and to identify the factors responsible for the persistence of MCPyV. In order to achieve this objective, an MCPyV 3D tissue infection model was established, which is based on a previously published IPSC-derived skin organoid model and which represents the first infection model for this human tumor virus. In this model, previously identified central molecular switches of viral infection will be analyzed.
We will focus on mutants of viral gene products such as sT, of ALTO and the viral miRNA, miR-M1, all of which were identified to contribute to viral persistence. In addition, we target with inhibitors or genetic manipulation important immune regulatory switches expressed by the host cells.
We will employ sequencing and advanced imaging tools developed within the RU DEEP-DV including single-cell RNA sequencing (scRNAseq), single-cell ATAC sequencing (scATACseq) and spatial transcriptomics.
This project is part of the DFG-funded research group FOR5200 DEEP-DV .
Our team members Silvia Albertini (PostDoc) and Lisann Röpke (PhD student) are dedicated to this project.
Structural characterization of the Merkel Cell Polyomavirus Tumor antigens
MCPyV-positive Merkel cell carcinomas (MCCs) are exclusively dependent on the expression of the MCPyV T antigens, small T (sT) and truncated large T antigen (trLT). In addition to tumorigenesis, the full-length LT protein functions as a helicase during viral replication. It initiates replication by binding to GRGGC motifs within the viral origin of replication (ori) via its origin binding domain (OBD). Both domains, helicase and OBD, are C-terminal and are partially or completely lost during tumorigenesis. We have recently shown that the MCPyV LT C-terminus (which is not present in MCC) contributes to genotoxic stress ( Theiss et al. ).
We speculate that functions of the C-terminal regions of LT may contribute to early events of MCC tumorigenesis, but may be dispensable or even detrimental for the fully transformed cancer cell phenotype and thus may be one of the factors exerting selective pressure for LT truncation. In this research project, we are using a structural approach to learn more about the structural and functional differences of the LT proteins encoded by polyomaviruses. In collaboration with the CSSB , we are performing single particle electron cryo-microscopy (cryoEM) of clinically relevant human polyomavirus LT proteins.
This project is part of the DFG-funded research group FOR5200 DEEP-DV .
Our team member Veronika Brinschwitz (PostDoc) is dedicated to this project.
Pathogenesis
Characterization and function of extracellular vesicles in Merkel Cell Cancer progression
Extracellular vesicles (EVs) have gained interest as intercellular communicators in various diseases, including infection and cancer. In cancer progression, EVs play a significant role in interactions within the tumor microenvironment (TME). However, the function of EVs in the pathogenesis of Merkel cell carcinoma (MCC) and concerning modifications and functions in the TME remains elusive. Individual publications show miRNA-375 as an EV cargo, which is transfered via EVs in fibroblasts and thus suggested to activate the TME. Our group showed that in MCPyV positive MCC tumors, the viral oncoprotein sT is crucial for tumor progression by altering surface marker expression and thereby TME interaction.
We performed a comprehensive description and analysis of the protein and nucleic acid cargos of EVs derived from patient-derived MCC cell lines. Further we analyze potential differences in cargo load in dependence of MCPyV sT protein. RNA and proteome analysis suggest that EV cargos do not simply reflect the parental cell composition. Notably, most highly expressed proteins, transcripts, and miRNAs from the parental cells were not detected in EVs. Interestingly, fragmented and intron-containing mRNAs were highly abundant in MCC-derived EVs suggesting that these RNAs are non functional as templates for protein expression. Small RNA Seq analysis confirm miR-375 as one of the most abundant miRNA cargos together with several other miRs involved in tumorigenesis. Proteome analysis shows an enrichment of histones, particularly in EVs derived from cells expressing sT protein. Based on this comprehensive characterization of MCC EVs, current experiments address the qualitative differences of EVs in MCC progression, and their functional role especially in activation of TME resident macrophages and immune evasion.
This project is part of the DFG-funded Research Training Group 2771 .
Our team member Ute Westerkamp (PhD student) is dedicated to this project.
Characterization of the MCPyV LT proteome
Merkel cell polyomavirus (MCPyV) is a small, double-stranded DNA virus that has been identified as the major causative agent of Merkel cell carcinoma (MCC), a rare but aggressive skin cancer. Central to MCPyV's oncogenic potential, particularly in the initial steps of tumorigenesis and possibly integration, is its large T (LT) antigen, a multifunctional protein that plays a critical role in viral replication. The LT antigen contains several conserved domains, including a DnaJ domain, a retinoblastoma protein (pRb) binding motif (LXCXE), a nuclear localization signal (NLS) and a helicase/ATPase domain, which together mediate interactions with host cellular machinery. These interactions disrupt normal cell cycle regulation, promote cell proliferation and contribute to the development of malignancy. In MCC tumors, the LT is often truncated, retaining the pRb-binding motif but lacking the C-terminal helicase domain, a feature that appears to be critical for oncogenesis while avoiding cytotoxicity. Understanding the MCPyV LT proteome in MCPyV target cells - both the full-length protein and the truncated LT protein when bound to viral or host DNA - is essential to elucidate the function of LT in infection and pathogenesis. In this study, we apply TurboID fused to LT protein (full length, truncated in the presence or absence of viral or host DNA) to biotinylate and subsequently identify all proteins in close proximity to the LT bait proteins in human dermal fibroblasts.
This project is part of the DFG-funded Graduate School RTG2887 VISION .
Our team member Tommaso Mari (PhD student) is dedicated to this project.
Antivirals
Antiviral mechanism of BK virus specific inhibitors
PyV-associated nephropathy (PVAN), caused by reactivated BKPyV in immunosuppressed hosts, reflects uncontrolled viral replication and subsequent tissue damage. Currently, no specific antiviral treatment is available and supportive care is limited to reducing immune suppression, which carries the risk of graft rejection. This deficiency is attributed to the lack of small animal models together with poor surrogate in vitro/in vivo systems, e.g. simian virus, SV40, which do not reflect the pathogenicity strategies of BKPyV. Specific features of BKPyV are its persistence in the kidney and its reactivation under immunosuppression. The mechanism of reactivation and the precise cell type of persistence are unknown. Depending on the cell type, BKPyV infection/reactivation results in massive viral replication, reassortment of viral variants and subsequent tissue invasion. Alternatively, BKV infection can result in prolonged viral protein expression, low viral particle yields and no visible cell lysis, for example in endothelial cells of vascular or urinary origin.
This project focuses on the following two challenges in BKPyV research: (1) lack of infection models that reflect BKPyV infection, e.g. virus entry and spread (2) lack of antiviral treatment to inhibit BKPyV reactivation and replication.
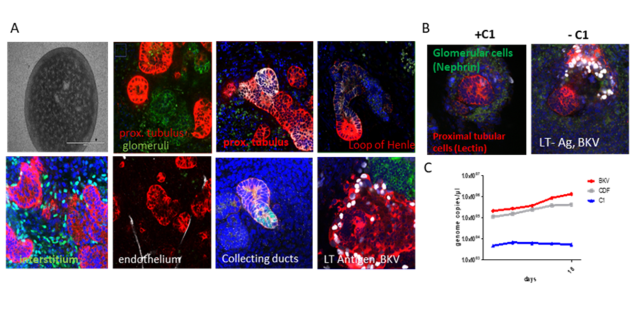
We recently established renal organoids differentiated from induced pluripotent stem cells (Fig.1A). We successfully show the correct differentiation of the different cell types in the organoid (Fig. 1B) and its successful infection (Fig. 1C). Furthermore, initial experiments show that these tissues are suitable to characterize BKV specific inhibitors (Fig. 1C).
Aspects of the project are part of the DZIF TTU IICH (kidney organoids as a preclinical model to text antivirals against BKPyV infection).
The project part focusing on characterizing the BKPyV life cycle in primary target cells using small molecule inhibitors is integrated in the DFG-funded Research Training Group 2771 .
Our team members Hannes Roggenkamp (clinical scientist) and Clara Husser (PhD student) are dedicated to these projects.
Preclinical 3D infection models for antiviral drug testing
In collaboration with our research partners, our scientific interest lies in the area of opportunistic infections in immunocompromised hosts. Our research focuses particularly on infections caused by herpes viruses (HSV-1/-2, VZV, EBV, KSHV) and polyomaviruses (JC, BK, MCPyV). These viruses are responsible for the development of life-threatening diseases, which present in a variety of ways and require the use of advanced infection models. These include skin for eczema herpeticum, skin/mucosa for genital herpes, brain for HSV-1/-2 and JCPyV, and kidney for BKPyV and PyVAN. In collaboration with our partners, we are developing 3D infection tissue models for these viruses and applying high-resolution microscopy, transcriptomics, and chromatin analysis at the single-cell level and with spatial resolution in these models. These models and methods will be of increasing interest in the future for the preclinical development phase of therapeutics against various pathogens.
Our team members Silvia Albertini (scientist) and Hannes Roggenkamp (clinical scientist) are dedicated to this DZIF TTU IICH project.
Identification and hit-to-lead development of BKPyV inhibiting compounds
Due to the complete lack of a specific anti-viral therapy, treatment options of BKPyV reactivation are mainly restricted to reconstitution of the immune system by alleviation of the immunosuppressive regimen. At the MVTV we aim to identify small molecule inhibitors active against BKPyV and their further development. The project has a clear translational focus in that it will further characterize the mode of action of a promising previously identified inhibitor of BKPyV infection. To date, there are no specific therapies, including antivirals, against BKPyV infection. The project is supported scientifically and strategically within the AiCuris AIcubator program.
This project is a joint collaboration between the LIV, research unit Virus Genomic and the MVTV anchored in the DZIF, TTU IICH .
Metagenomics
Developing pre-analytical and bioinformatic tools to detect and identify pathogens in clinical samples.
Together with partners at the Leibniz Institute for Virology ( LIV - Research Unit Virus Genomics ), we have established a Next Generation Sequencing (NGS) platform, which allows hypothesis-free detection of known as well as unknown/new infectious agents in clinical samples.
In particular, this includes the development of bioinformatics software (DAMIAN) that performs complete data processing in an automated manner to determine the microorganisms present ( Alawi et al. 2019 , de Vries et al. 2021 , de Vries et al. 2021 , Lopez-Labrador et al. 2021 ). Using this software, we have been able to detect various pathogens in clinical samples and identify several new viruses ( Becher et al. 2014 , Fischer et al. 2014 , Neill et al. 2014 , Baechlein et al. 2015 , Corman et al. 2015 , Postel et al. 2015 , Baechlein et al. 2016 , Postel et al. 2016 , Postel et al. 2016 , Brancaccio et al. 2017 , Gunther et al. 2017 , Postel et al. 2017 , Fux et al. 2019 , Rubbenstroth et al. 2019 ).
As a result of the previous implementation of our analysis workflows, we have been coordinating the SARS-CoV-2 Genome Surveillance in Hamburg, funded by the Hanseatic City of Hamburg. From an early stage of the pandemics (since March 2020), we were able to perform whole-genome sequencing of SARS‐CoV‐2 in clinical samples. We were able to track the emergence of mutations and resolve transmission chains in Hamburg and a large outbreak in a German meat processing plant in Gütersloh during the summer of 2020 ( Gunther et al. 2020 , Pfefferle et al. 2020 , Pfefferle et al. 2021 ). From March 2020 to March 2023, we thus far have sequenced more than 15.000 full-length genomes. We sequenced approximately 150‐200 cases per week and provided a weekly report of our analysis to authorities and the general public.
Benchmarking of metagenomics in clinical samples
Within this implementation of NGS in diagnostics, we are in close exchange with other international working groups to standardize the application of metagenomics in clinical samples and work on accreditation concepts. In particular, we have a close exchange with the European Society for Clinical Virology (ESCV) to develop standardized procedures and perform inter-laboratory comparisons ( de Vries et al. 2021 , de Vries et al. 2021 , Lopez-Labrador et al. 2021 ).
Metagenomics in emerging viral infections – integrating non-host and host transcriptional responses to detect and predict viral infection
Viral infections pose a major challenge to our healthcare system. Such new emerging infections must therefore be detected at an early stage and their impact adequately assessed. Although our current diagnostics are well advanced, we still have difficulties in identifying infectious agents, especially novel or highly diverse ones. Making a diagnosis remains inadequate, especially very early in infection or when the pathogen is not present in non-invasive diagnostic specimens. This has consequences for the infection process at the community level, while delaying appropriate therapy at the patient level. To identify infectious agents, even when there are low pathogen loads, we will use metagenomic analyses for virus/pathogen detection in combination with host response assessment. The project will build on existing expertise in viral genomics and metagenomics for the detection of viral, bacterial, and other non-host sequences, address the existing gaps in current microbial diagnostics for the detection of potential pathogens and develop disease-associated host responses for the prediction of infectious diseases. In particular, the project addresses a prospective approach for detecting the unknown pathogen X for its early detection and implementation of countermeasures.
This project is funded within DFG's Collaborative Research Center 1638 .
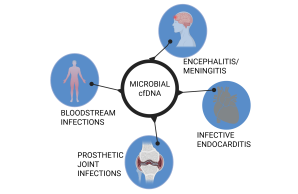
mcf-DNA sequencing for the diagnosis of pathogens in plasma.
Timely initiation of antimicrobial therapy is a critical step in preventing morbidity and mortality associated with infection. Diagnosis of infection is often difficult, especially in the immunocompromised host, due to non-classical symptoms and a broad spectrum of pathogens. Culture-independent methods have advantages, but are also challenging in terms of sensitivity. Molecular diagnostics combined with liquid biopsy as a non-invasive sample source, especially in immunocompromised and pediatric patient groups, represent a promising diagnostic approach. Originally developed in the field of oncology for the early detection of circulating tumor cells, cell-free DNA (cf-DNA) sequencing can also be used for the detection of pathogens in plasma. Microbial cell-free DNA (mcf-DNA) is based on fragments of pathogen DNA shed into the bloodstream from a distant source of infection. It allows identification of species including bacterial, viral (DNA viruses), fungal or parasitic organisms. First studies have shown up to 90% sensitivity in detecting mcf-DNA from plasma of patients with sepsis and acute endocarditis. Mcf-DNA Seq can also be useful for identifying occult infections in a variety of clinical scenarios. Within this project we work in close collaboration with other DZIF members to develop protocols for preanalytical and bioinformatic processing of plasma samples to isolate and sequence cfDNA with the subsequent identification of pathogens.
This project is closely related to the DZIF Bridging Topic Diagnostics .
Diagnostic application
Several of these methods will be implemented in diagnostic applications. This application in diagnostic samples is based on an infrastructure that has been continuously developed over several years between the UKE and the LIV.
_contentbild2.png)