Work Group "Network Neuroscience"
- Brain network architecture
- Modelling brain network dynamics
- Brain network pertubation
-
Brain network architecture
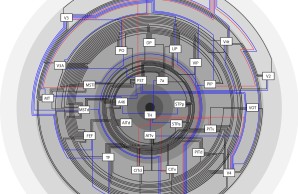
Brain network architecture
How are brain networks organized? We apply multivariate and graph-theoretical analyses to extensive databases of inter-regional connections in the mammalian brain. Our studies of model species such as the rhesus macaque, cat and mouse have shown that structural brain networks possess a highly modular and hierarchical organization. Moreover, short communication paths appear to be an important constraint for the organization of neural networks across several dimensions of scale. Together with our collaborators we have also explored fundamental relations between the organization of brain architecture and brain connectivity, identifying as a general principle across mammalian species that the existence and laminar origin and termination patterns of connections are strongly linked to the the architectonic similarity of cortical areas.
Research Topics
- building a database of ferret brain connectivity and architecture (the 'ferretome')
- multivariate analyses of brain architecture and network organization
- exploring structural and functional constraints on brain networks
- simulating brain network development and evolution
Key Publications
Hilgetag CC, Amunts K (2016) Connectivity and cortical architecture, e-Neuroforum 7: 56
Hilgetag CC, Medalla M, Beul S, Barbas H (2016) The primate connectome in context: principles of connections of the cortical visual system, NeuroImage 134: 685
Sukhinin DI, Engel AK, Manger P, Hilgetag CC (2016) Building the Ferretome, Front Neuroinf 10:16
Goulas A, Uylings HBM, Hilgetag CC (2016) Principles of ipsilateral and contralateral cortico-cortical connectivity in the mouse, Brain Struct Funct, DOI: 10.1007/s00429-016-1277-y
Hilgetag CC, Goulas A (2016) Is the brain really a small-world network? Brain Struct Funct 221: 2361
Beul S, Grant S, Hilgetag CC (2015) A predictive model of the cat cortical connectome based on cytoarchitecture and distance, Brain Struct Funct 220: 3167
Hilgetag CC, Grant S (2010) Cytoarchitectural differences are a key determinant of laminar projection origins in the visual cortex, NeuroImage 51: 1006
-
-
Modelling brain network dynamics
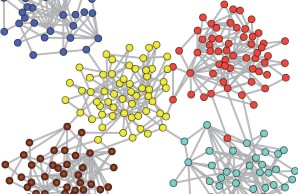
Modeling brain network dynamics
We employ computational models of different complexity in order to understand the influence of topological features of brain connectivity on brain dynamics. The modelling findings suggests that the hub and modular features of brain networks are reflected in their spatial-temporal activation patterns, and that the hierarchical arrangement of multi-scale cortical networks supports stable functioning for a wide range of activation parameters. Such computational models can also give hints on pathological brain dynamics in conditions such as epilepsy or after stroke.
Research Topics
- exploring the influence of network topology on network dynamics using models of different complexity (excitable dynamics, neural mass models, multi-scale models)
- modeling the contribution of extrinsic and intrinsic connections in laminar cortical circuits
Key Publications
Messe A, Hütt MT, König P, Hilgetag CC (2015) A closer look at the apparent correlation of structural and functional connectivity in excitable networks, Scientific Reports 5: 7870
Garcia GC, Lesne A, Hilgetag CC, Hütt MT (2014) Role of long cycles in excitable dynamics on graphs, Phys Rev E 90: 052805
Hütt MT, Kaiser M, Hilgetag CC (2014) Network-guided pattern formation of neural dynamics. Phil Trans R Soc B 369, 20130522
Hilgetag CC, Hütt MT (2014) Hierarchical modular brain connectivity is a stretch for criticality, Trends Cogn Sci 18: 114
Garcia GC, Lesne A, Hütt MT, Hilgetag CC (2012) Building blocks of self-sustained activity in a simple deterministic model of excitable neural networks, Front Comput Neurosci 6:50.
Müller-Linow M, Hilgetag CC, Hütt MT: Organization of excitable dynamics in hierarchical biological networks, PLoS Comput Biol 4 (2008) e1000190.
Kaiser M, Goerner M, Hilgetag CC (2007) Criticality of spreading dynamics in hierarchical cluster networks without inhibition, New J Phys 9: 110.
Zhou C, Zemanova L, Zamora G, Hilgetag CC, Kurths J (2006) Hierarchical organization unveiled by functional connectivity in complex brain networks, Phys Rev Lett, 268103 (2006)
-
-
Brain network pertubation
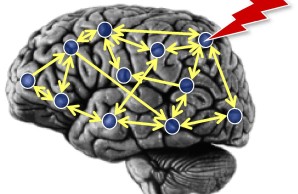
Brain network perturbation
To probe the causal functional contributions of different features of brain networks to behavior, we use the experimental technique of transcranial magnetic stimulation (TMS) as well as theoretical approaches for lesion inferences. In particular, a game-theoretical approach for the analysis of multiple brain perturbations provides a helpful perspective for neurological lesion inference as well as for targeting therapeutic interventions in brain-damaged patients.
Research Topics
- non-invasive perturbation of the human brain using TMS
- lesion inferences of brain damage and perturbations in animal models and the human brain
Key Publications
Zavaglia M, Forkert ND, Cheng B, Gerloff C, Thomalla G, Hilgetag CC (2015) Mapping causal functional contributions derived from the clinical assessment of brain damage after stroke, NeuroImage:Clinical 9: 83
Zavaglia M, Hilgetag CC (2015) Causal functional contributions and interactions in the attention network of the brain: An objective multi-perturbation analysis, Brain Struct Funct 221: 2553
Rushmore RJ, Valero-Cabre, A, Lomber SG, Hilgetag, CC, Payne BR (2006) Functional circuitry underlying visual neglect, Brain 129: 1803
Keinan A, Sandbank B, Hilgetag CC, Meilijson I, Ruppin E (2004) Fair attribution of functional contribution in artificial and biological networks, Neural Computation 16: 1887
Hilgetag CC, Théoret H, Pascual-Leone A (2001) Enhanced visual spatial attention ipsilateral to rTMS-induced ‘virtual lesions’ of human parietal cortex, Nature Neuroscience 4: 953
Young MP, Hilgetag CC, Scannell JW (2000) On imputing function to structure from the behavioural effects of brain lesions, Phil Trans R Soc Lond B 355: 147
-
Work Group "Image Processing and Medical Informatics"
- Advanced Biomedical Image Computing
- Image-based Analysis and Modeling
- eLearning
-
Advanced Biomedical Image Computing
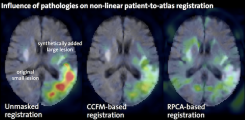
Advanced Biomedical Image Computing
How can we reliably and efficiently extract meaningful information from medical and biomedical image data?
Medical and biomedical image data plays an increasingly important role in clinical routine and research. Novel imaging modalities and protocols, increased image resolution and highly application-dependent demands require novel targeted and efficient image processing solutions and pipelines.
Together with our collaborators we develop such targeted advanced (neuro-)image processing techniques and solutions. Our research focus is medical image computing under challenging conditions and beyond capabilities of standard toolkits:
- Development and optimization of problem-specific non-linear image registration approaches
- Processing of artifact-affected data and atlas-based image analyses in the presence of pathologies
- Integrating machine learning approaches into established medical image analysis frameworks
Collaboration Partners
- Institute for Biomedical Imaging, Hamburg University of Technology
- Siemens Healthcare GmbH
- Department of Radiotherapy and Radio-Oncology, UKE
- Department of Radiology & Hotchkiss Brain Institute, University of Calgary
Key Publications
Werner R, Wilms M, Cheng B, Forkert ND (2016) Beyond cost function masking: RPCA-based non-linear registration in the context of VLSM. In: 2016 International Workshop on Pattern Recognition in Neuroimaging (PRNI), Trento, Italy; IEEE Xplore, pp. 1-4, 2016.
Werner F, Jung C, Hofmann M, Werner R, Salamon J, Säring D, Kaul MG, Them K, Weber OM, Mummert T, Adam G, Ittrich H, KnoppT (2016) Geometry planning and image registration in Magnetic Particle Imaging using bimodal fiducial markers, Med Phys 43: 2884-93
Werner R, Schmidt-Richberg A, Handels H, Ehrhardt J (2014) Estimation of lung motion fields in 4D CT data by variational non-linear intensity-based registration: A comparison and evaluation study, Phys Med Biol 59: 4247-60
-
-
Image-based Analysis and Modeling
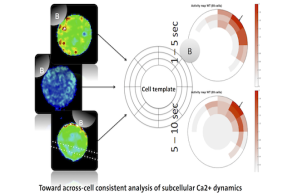
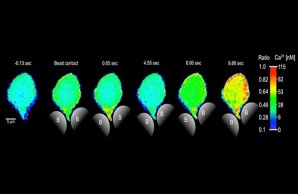
Image-based Analysis and Modeling of Dynamic Processes
One of the most challenging fields of biomedical image computing is the analysis of spatio-temporal image series and image-based modeling of the represented dynamic processes.Adapting and extending non-linear registration methods, classic multi-variate statistical analysis techniques and machine learning approaches, we address processes on diverse temporal and spatial scales, ranging from, e.g., subcellular Ca2+ dynamics in T cells to pulsatile deformation of the cerebral vasculature.
Collaboration Partners
- Department of Biochemistry and Molecular Cell Biology, UKE
- Institute of Medical Technology, Hamburg University of Technology
- Neuroradiological Diagnostics and Intervention, UKE
Selected Publications
Schetelig D, Säring D, Illies T, Sedlacik J, Kording F, Werner R (2016) Phantom-based ground-truth generation for vessel segmentation and pulsatile deformation analysis. In: Webster J, Yaniv ZR (eds.), SPIE Medical Imaging: Image-Guided Procedures, Robotic Interventions, and Modeling, San Diego, USA; SPIE Proc. 9786, pp. 978622-1-6.
Wolf IMA, Diercks B-P, Gattkowski E, Czarniak F, Kempski J, Werner R, Schetelig D, Mittrücker H-W, Schumacher C, von Osten M, Lodygin D, Flügel A, Fliegert R, Guse AH (2015) Frontrunners of T cell activation: Initial, localized Ca2+ signals mediated by NAADP and the type 1 ryanodine receptor, Sci Signal 8:ra120
Tehrani J, Yang Y, Werner R, Lu W, Low D, Guo X, Wang J (2015) Sensitivity of tumor motion simulation accuracy to lung biomechanical modeling approaches and parameters, Phys Med Biol 60:8833-49
Wilms M, Werner R, Ehrhardt J, Schmidt-Richberg A, Schlemmer HP, Handels H (2014) Multivariate regression approaches for surrogate-based diffeomorphic estimation of respiratory motion in radiation therapy, Phys Med Biol 59: 1147-64, 2014
Marx M, Ehrhardt J, Werner R, Schlemmer HP, Handels H (2014) Simulation of spatio-temporal CT data sets using a 4D MRI-based lung motion model, Int J Comput Assist Radiol Surg 9: 401-9
Polzin T, Rühaak J, Werner R, Handels H, Modersitzki J (2014) Lung registration using automatically detected landmarks, Methods Inform Med 53: 250-6
-
-
eLearning

eLearning
The UKE eLearning platform Mephisto/UKE based on the open source platform Moodle has been installed by a team under the supervision of the predecessor of our Image Processing and Medical Informatics group and funded by the Förderfond Lehre of the University of Hamburg Medical Faculty. Meanwhile, Mephisto and extensions like iMED textbook and room administration together with a comfortable iMED student management system offer a central tool for the medical education at the UKE.
Mephisto/UKE not only provides teaching materials and continuous communication between teachers and students, but also subject-specific online links to allow our medical students to efficiently study in- and outside classrooms as well as the opportunity to integrate interactive multimedia-based content like virtual patients, virtual microscopy, test exams, application-specific software simulators and courses together with more teaching methods including blended learning. It also allows booking facilities for limited resources like course participation or student contributions.
Within this context, our current research includes investigating the effective use and creation of e.g. virtual patients, virtual microscopy, virtual emergency rooms and more. This extends the (e)Learning opportunities at the UKE.
Main Collaboration Partners
- III. Department of Internal Medicine, University Medical Center Hamburg-Eppendorf (S. Harendza)
- Department of Anatomy and Experimental Morphology, University Medical Center Hamburg-Eppendorf (G.H. Lüers)
- Department of Biochemistry and Molecular Cell-Biology, University Medical Center Hamburg-Eppendorf (W. Hampe)
- Department of General and Interventional Cardiology, University Heart Center Hamburg, University Medical Center Hamburg-Eppendorf (B.-Chr. Zyriax)
Key Publications
Jäger F, Riemer M, Abendroth M, Sehner S, Harendza S (2014) Virtual patients: the influence of case design and teamwork on students’ perception and knowledge – a pilot study BMC Med Educ 14:137
Abendroth M, Harendza S, Riemer M (2013) Clinical decision making: A pilot e-learning study Clin Teach 10: 51-5
Riemer M, Hampe W, Wollatz M, Peimann C-J, Handels H (2007) Erste Erfahrungen mit der eLearning-Plattform Moodle im Universitätsklinikum Hamburg-Eppendorf -Evaluationsergebnisse im Querschnittsfach Medizinische Informatik und der Biochemie In: Kundt G, et al. (Hrsg.), eLearning in der Medizin, Proceedings, CBT 2007, pp. 235-246
-