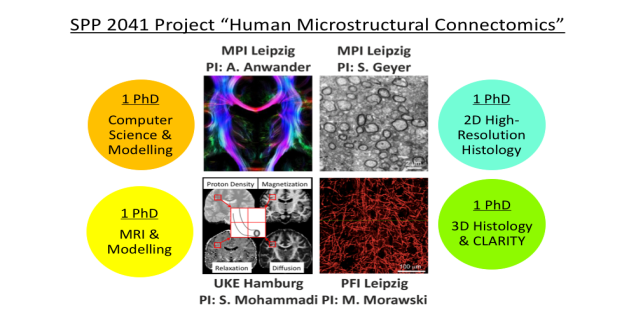
In this project four PhD students will work closely together on developing models and techniques to create a better map of the human microstructural connectome, in vivo and ex vivo using MRI and histology.
Long-range connections comprise only about 10% of total CNS connections. Their arrangement, length, and microstructural properties are of utmost importance for the functional organization of the CNS, because they determine how information is distributed across the brain. In particular, axon diameter and g-ratio (the ratio between axonal body and axon plus myelin sheath) is related to the conduction velocity and thus relevant for the information transfer rate in a functional CNS network. To date, diffusion magnetic resonance imaging (dMRI)-based tractography is the only in vivo technique for mapping the structural long-range connections in the human brain. However, mapping from diffusion to fiber pathways is still ill-posed. Tractography algorithms can take "wrong turns" and produce a number of false connections.
To address this significant limitation, microstructure-informed tractography has been suggested. Microstructure-informed tractography is an emerging computational framework that associates each computed fiber tract with microstructural properties, e.g., metrics for axon diameter or density, using the dMRI technique. However, in its current form microstructure-informed tractography suffers from three major problems:
- (i) to estimate these metrics biophysical models are used, which in turn aim at solving another ill-posed task: relate the macroscopic MR signal to microscopic tissue properties,
- (ii) to stabilize the ill-posed biophysical model a lower spatial resolution is required and thus coarse spatial resolution is applied thereby increasing the number of voxels with ambiguous fiber configurations,
- (iii) it relies on the untested neuroanatomical hypothesis that two adjacent fiber pathways show less within-fiber than across-fiber variation.
In this highly inter-disciplinary project, we will develop a computational framework for microstructure-informed tractography that addresses the aforementioned limitations using multi-modal quantitative MRI at an unprecedented ultra-high spatial resolution (660 microns). Moreover, we will develop an advanced ex vivo histology analysis strategy, using complementary 2-D (high-resolution semithin and ultrathin sectioning) and 3-D (CLARITY) techniques. We will fuse gold-standard ex vivo histology with MRI to validate the proposed model at central junctions of long-range fiber pathways within the well characterized human voluntary motor control network. By emphasizing the close integration of multi-modal computational biophysical models, advanced MRI technology (the German-wide unique combination of a CONNECTOM & 7T MRI system), and advanced histological approaches (CLARITY in human tissue), this project aims at a paradigm shift in in vivo MRI-based computational models for in vivo tractography.
Depending on where you find yourself in this project (see Figure), you can directly contact the respective PI to learn more about the project (see also our project website ):
Alfred Anwander:
anwander@cbs.mpg.de
Stefan Geyer:
sgeyer@cbs.mpg.de
Siawoosh Mohammadi:
s.mohammadi@uke.de
Markus Morawski:
Markus.Morawski@medizin.uni-leipzig.de