P5 - Survival prediction of prostate cancer patients from histopathology images and electronic health records
Stefan Bonn & Marina Zimmermann
Institute for Medical Systems Biology (IMSB), University Medical Center Hamburg-Eppendorf
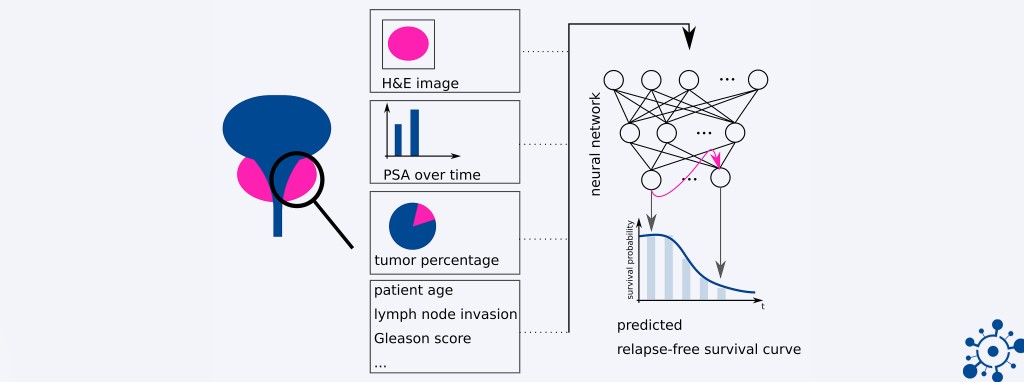
At the IMSB we aim to build AI-based clinical decision support systems for Prostate Cancer (PCa) with our collaboration partners at the Department of Pathology at the UKE and the Martini-Klinik . Currently, the diagnosis of PCa and the treatment decision are based on Gleason scores and on nomograms. However, Gleason scores suffer from high inter-observer variability and nomograms are only able to model linear dependencies between patient parameters. Therefore, the goal of our research is to provide individual and objective prognoses for PCa patients through predicting relapse after radical prostatectomy (RPE). To this end we build and train deep learning-based models that predict the probability of a patient having a relapse from either H&E-stained images of biopsies after the RPE or electronic health records that reflect the patient’s clinical history related to prostate cancer. We can show that our survival models perform on par or outperform predictions based on current clinical scores and have the advantage of showing the course of the probability of relapse over time.
Partner: G. Sauter, M. Lennartz, Institute for Pathology, UKE; L. Budäus, P. Tennstedt, M. Kachanov, Martini-Klinik
Contact: sbonn@uke.de
-
Selected publications on topic
Towards Explainable End-to-End Prostate Cancer Relapse Prediction from H&E Images Combining Self-Attention Multiple Instance Learning with a Recurrent Neural Network. Dietrich E, Fuhlert P, Ernst A, Sauter G, Lennartz E, Stiehl S, Zimmermann M, Bonn S, Machine Learning for Health (ML4H) 2021. Cornell University, 38-53.
Deep Learning-Based Discrete Calibrated Survival Prediction. Fuhlert P, Ernst A, Dietrich E, Westhaeusser F, Kloiber K, Bonn S. IEEE International Conference on Digital Health (ICDH) 2022, 169-174.