

Tools and techniques to establish tailored assays to study the biology of emerging viruses and identify their Achilles’ heels
Summary
This service project provides advanced technologies that help researchers within the CRC study how viruses interact with human cells. These tools include genetic modification techniques, pseudoviruses for safe experiments, and CRISPR/Cas genome editing. The goal is to support various research groups with tailored methods to better understand virus biology and immune responses.
One key focus is the use of lentiviral vectors - engineered viruses that can deliver genes into cells. These vectors are adapted to different research needs, such as testing how viruses enter cells, finding new receptors, or identifying neutralizing antibodies. In the future, the project will also help identify specific virus fragments (epitopes) that are recognized by T cells, a crucial part of the immune system.
Since the project is designed to support many partners, its work will evolve based on the needs of each individual research project within the CRC.
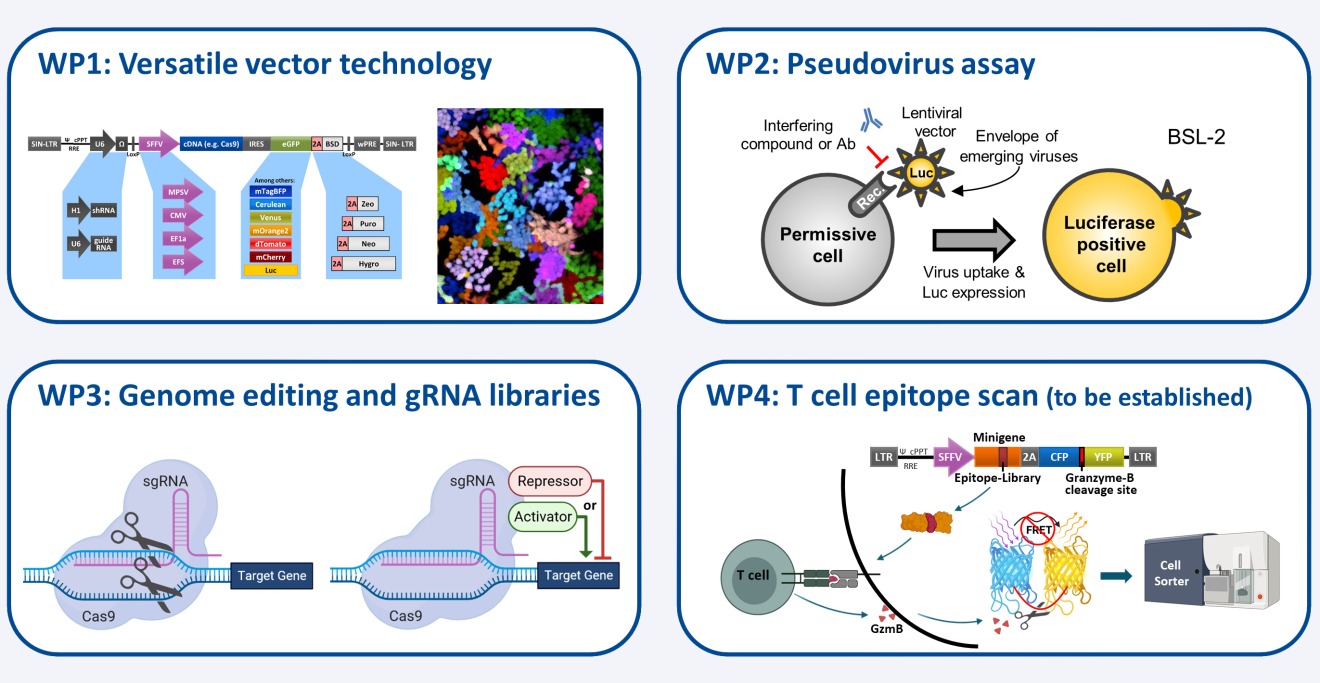
Our Work Packages (WP):
WP1: Genetic modification of human and mouse cells - we provide customized lentiviral vectors for stable gene expression or knockdown, tailored to different cell types and research goals.
WP2: Pseudoviruses to study virus entry and immune responses - we create safe-to-use pseudotyped viral particles to analyze how viruses infect cells and how antibodies block this process.
WP3: CRISPR/Cas-based gene editing and screening - we offer tools for precise genome editing and genome-wide CRISPR screens to identify host genes involved in virus infection.
WP4: Tools to identify T-cell epitopes - we are developing a new method to find virus-specific protein fragments that are recognized by T cells, supporting vaccine and immunology research.
These work packages help us to provide flexible and powerful tools that support CRC researchers in modifying cells, studying virus entry, identifying host factors, and uncovering immune targets - all essential steps in understanding and combating emerging viral diseases.
Team


Prof. Dr. Boris Fehse
Principal Investiagor
E-mail address:
Phone: +49 (0) 40 7410 - 55250


Dr. Kristoffer Riecken
Principal Investigator
E-mail adress:
Phone: +49 (0) 40 7410 - 52705


Research Department Cell and Gene Therapy
The Research Department Cell and Gene Therapy, a part of the Department of Stem Cell Transplantation, is based at the University Medical Center Hamburg-Eppendorf (UKE).
Project related publications
Winterberg KJ#, Möller C#, Schwarze LI, Cichutek S, Hambach J, Guse AH, Fehse B, Diercks BP#, Riecken K#. Imaging BiTE-Mediated Activation of Primary Human CD8+ T Cells. Methods Mol Biol. 2025;2904:159-177. doi: 10.1007/978-1-0716-4414-0_12.
Seifert L#, Riecken K#, Zahner G, Hambach J, Hagenstein J, Dubberke G, Huber TB, Koch-Nolte F, Fehse B, Tomas NM. An antigen-specific chimeric autoantibody receptor (CAAR) NK cell strategy for the elimination of anti-PLA2R1 and anti-THSD7A antibody-secreting cells. Kidney Int. 2024 Apr;105(4):886-889. doi: 10.1016/j.kint.2024.01.021.
Enk LUB#, Hellmig M#, Riecken K, Kilian C, Datlinger P, Jauch-Speer SL, Neben T, Sultana Z, Sivayoganathan V, Borchers A, Paust HJ, Zhao Y, Asada N, Liu S, Agalioti T, Pelczar P, Wiech T, Bock C, Huber TB, Huber S, Bonn S, Gagliani N, Fehse B, Panzer U, Krebs CF. Targeting T cell plasticity in kidney and gut inflammation by pooled single-cell CRISPR screening. Sci Immunol. 2024 Jun 14;9(96):eadd6774. doi: 10.1126/sciimmunol.add6774.
Tintelnot J, Xu Y, Lesker TR, Schönlein M, Konczalla L, Giannou AD, Pelczar P, Kylies D, Puelles VG, Bielecka AA, Peschka M, Cortesi F, Riecken K, Jung M, Amend L, Bröring TS, Trajkovic-Arsic M, Siveke JT, Renné T, Zhang D, Boeck S, Strowig T, Uzunoglu FG, Güngör C, Stein A, Izbicki JR, Bokemeyer C, Sinn M, Kimmelman AC, Huber S#, Gagliani N#. Microbiota-derived 3-IAA influences chemotherapy efficacy in pancreatic cancer. Nature 2023;615:168-174. doi: 10.1038/s41586-023-05728-y.
Kuzikov M, Woens J, Zaliani A, Hambach J, Eden T, Fehse B, Ellinger B#, Riecken K#. High-throughput drug screening allowed identification of entry inhibitors specifically targeting different routes of SARS-CoV-2 Delta and Omicron/BA.1. Biomed Pharmacother. 2022;151:113104. doi: 10.1016/j.biopha.2022.113104.
Hagemann K, Riecken K, Jung J, Hildebrandt H, Menzel S, Bunders MJ, Fehse B, Koch-Nolte F, Heinrich F, Peine S, Schulze zur Wiesch J, Brehm TT, Addo MM, Lütgehetmann M, Altfeld M. Natural killer cell-mediated ADCC in SARS-CoV-2-infected individuals and vaccine recipients. Eur J Immunol. 2022;52:1297-1307. doi: 10.1002/eji.202149470.
Badbaran A, Mailer RK, Dahlke C, Woens J, Fathi A, Mellinghoff SC, Renné T, Addo MM, Riecken K, Fehse B. Digital PCR to quantify ChAdOx1 nCoV-19 copies in blood and tissues. Mol Ther Methods Clin Dev. 2021;23:418-23. doi: 10.1016/j.omtm.2021.10.002.
Giersch K, Bhadra OD, Volz T, Allweiss L, Riecken K, Fehse B, Lohse AW, Petersen J, Sureau C, Urban S, Dandri M, Lütgehetmann M. Hepatitis delta virus persists during liver regeneration and is amplified through cell division both in vitro and in vivo. Gut 2019;68:150–57. doi: 10.1136/gutjnl-2017-314713.
Ellinger B, Pohlmann D, Woens J, Jäkel FM, Reinshagen J, Stocking C, Prassolov VS, Fehse B#, Riecken K#. A High-Throughput HIV-1 Drug Screening Platform, Based on Lentiviral Vectors and Compatible with Biosafety Level-1. Viruses 2020;12:E580. doi: 10.3390/v12050580.
Mock U, Machowicz R, Hauber I, Horn S, Abramowski P, Berdien B, Hauber J, Fehse B. mRNA transfection of a novel TAL effector nuclease (TALEN) facilitates efficient knockout of HIV co-receptor CCR5. Nucleic Acids Res. 2015;43:5560-71. doi: 10.1093/nar/gkv469.
Mohme M#, Maire CL#, Riecken K#, Zapf S, Aranyossy T, Westphal M, Lamszus K#, Fehse B#. Optical barcoding for single-clone tracking to study tumor heterogeneity. Mol Ther. 2017;25:621-33. doi: 10.1016/j.ymthe.2016.12.014.
Weber K’, Thomaschewski M, Warlich M, Volz T, Cornils K, Niebuhr B, Täger M, Lütgehetmann M, Pollok JM, Stocking C, Dandri M, Benten D, Fehse B. RGB marking facilitates multi-color clonal cell tracking. Nat Med. 2011;17:504-9. doi: 10.1038/nm.2338. (’changed name to Riecken)
Weber K’, Bartsch U, Stocking C, Fehse B. A multi-color panel of novel lentiviral "gene ontology" (LeGO) vectors for functional gene analysis. Mol Ther. 2008;16:698–706. doi: 10.1038/mt.2008.6. (’changed name to Riecken)
#equally contributing authors