
Project - A06:
The roles of short expressed peptides and cell type composition in autoimmune liver inflammation
Project leaders:
Prof. Dr. med. Norbert Hübner &
Prof. Dr. med. Christoph Schramm


Summary
Chronic damage to the liver such as immune-mediated and autoimmune inflammation results in excessive extracellular matrix production with consequential fibrosis and, in many cases, ultimately leads to cirrhosis. The mechanisms of uncontrolled inflammation and progressive fibrosis in the autoimmune liver diseases (AILD) primary sclerosing cholangitis (PSC) and autoimmune hepatitis (AIH) remain incompletely understood.
To gain new insights into the pathogenesis of AILD, we will study human hepatic translational events by measuring genome-wide active translation with ribosome profiling (Ribo-seq). This will allow us to identify novel short open reading frames (sORFs)-encoded peptides (SEPs) in RNA regions previously classified as non-coding (ncRNA). By comparing patients and controls, we will be able to detect disease-specific expression of annotated and novel peptides that may lead to the creation of new self-antigens, breaking of self-tolerance, triggering of autoimmune B and T cell responses, and / or modulation of inflammation or fibrosis. Since novel SEPs may have specialised roles in the function of specific cell types, knowledge about their expression across specific cell types will aid in the identification of their functional relevance. To address this, we will characterise hepatic cell types and states, expression networks, cellular circuits and spatial location by single-cell and spatial profiling approaches.
Moreover, by building a single-cell atlas for AILD, we will discern fundamental causes of maladaptive hepatic immune regulation, characterise heterogeneous cellular responses and infer new strategies to limit them. The analysis will allow us to identify and characterise rare subpopulations of intrahepatic cells that represent as little as 1% of the main population and harbour potential importance for liver homeostasis and disease development. Furthermore, it will unravel the heterogeneity, composition, potential function and spatial location of hepatic parenchymal and non-parenchymal subpopulations in AILD patients. The analysis of biopsy samples from an early stage of AILD in comparison to late stages will provide insight into initiating factors of the diseases and will show how fibrosis develops and progresses over time. Concurrently, computational tools for single-cell data analysis have been developed to follow dynamic processes such as cell differentiation and activation as well as explore interactions between cell populations based on their repertoire of ligands and receptors. While the emphasis of our studies will lie on PSC, the CRC network will enable us to extend our analyses to other AILD as a next step and various liver diseases in the future.
Project plan
We hypothesise that novel unannotated SEP are generated in chronically inflamed liver tissue of AILD patients that i) will have a modulatory effect on the disease process and ii) may expand the immunopeptidome in AILD. Moreover, we hypothesise that hepatic remodeling differs for each AILD etiology and correlates to changes in cell subpopulation prevalence and in cell type-specific gene expression. The molecular and cellular drivers underlying autoimmune and immune-mediated chronic liver inflammation and fibrosis remain incompletely understood. This project will focus on the identification of new AILD-specific SEPs. Moreover, we will define the cellular and transcriptional heterogeneity of AILD with single-cell sequencing approaches. Our long-term goal is to understand the function of novel disease-specific SEPs and to investigate the functional relevance of cell type-specific changes in AILD.
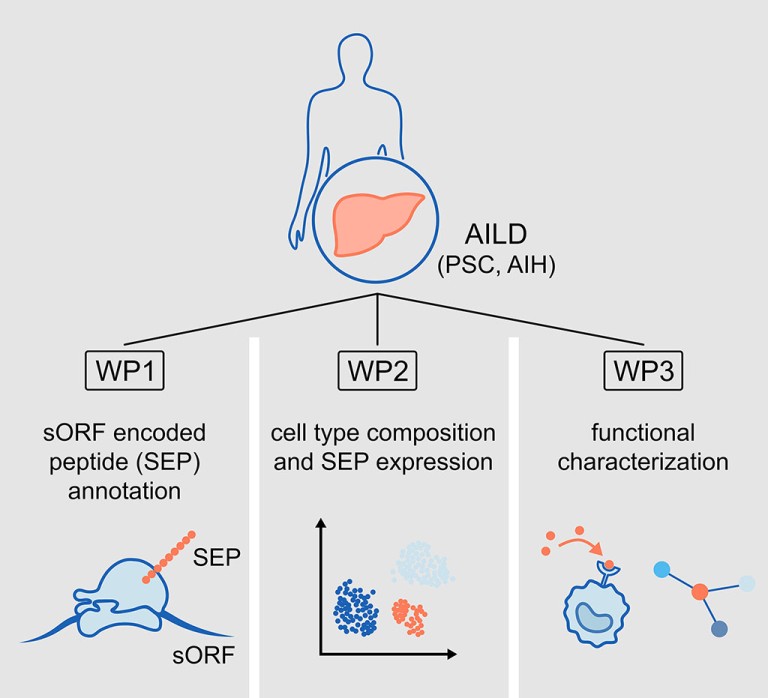
In order to test this hypothesis, our work programme has the following work packages (WP):
WP1: To detect, to validate and to characterise short ORF-encoded peptides (SEPs) in AILD.
WP2: To define hepatic cell type composition and SEP-expression in AILD.
WP3: To characterise candidate SEPs and their role in the immunopeptidome, cell function and interaction.
Project related publications
Van Heesch S, Witte F, Schneider-Lunitz V, Schulz JF, Adami E, Faber AB, Kirchner M, Maatz H, Blachut S, Sandmann CL, Kanda M, Worth CL, Schafer S, Calviello L, Merriott R, Patone G, Hummel O, Wyler E, Obermayer B, Mücke MB, Lindberg EL, Trnka F, Memczak S, Schilling M, Felkin LE, Barton PJR, Quaife NM, Vanezis K, Diecke S, Mukai M, Mah N, Oh SJ, Kurtz A, Schramm C, Schwinge D, Sebode M, Harakalova M, Asselbergs FW, Vink A, Weger RA de, Viswanathan S, Widjaja AA, Gärtner-Rommel A, Milting H, Remedios C dos, Knosalla C, Mertins P, Landthaler M, Vingron M, Linke WA, Seidman JG, Seidman CE, Rajewsky N, Ohler U, Cook SA, Hubner N. The Translational Landscape of the Human Heart. Cell 2019;178:242-260.e29. doi: 10.1016/j.cell.2019.05.010.
Chothani S, Schäfer S, Adami E, Viswanathan S, Widjaja AA, Langley SR, Tan J, Wang M, Quaife NM, Pua CJ, D’Agostino G, Shekeran SG, George BL, Lim S, Cao EY, Van Heesch S, Witte F, Felkin LE, Christodoulou EG, Dong J, Blachut S, Patone G, Barton PJR, Hubner N, Cook SA, Rackham OJL. Widespread Translational Control of Fibrosis in the Human Heart by RNA-Binding Proteins. Circulation 2019;140:937–51.doi: 10.1161/CIRCULATIONAHA.119.039596.
Schafer S, Adami E, Heinig M, Rodrigues KEC, Kreuchwig F, Silhavy J, Van Heesch S, Simaite D, Rajewsky N, Cuppen E, Pravenec M, Vingron M, Cook SA, Hubner N. Translational regulation shapes the molecular landscape of complex disease phenotypes. Nat Commun 2015;6:7200. doi: 10.1038/ncomms8200.
Sandmann CL, Schulz JF, Ruiz-Orera J, Kirchner M, Ziehm M, Adami E, Marczenke M, Christ A, Liebe N, Greiner J, Schoenenberger A, Muecke MB, Liang N, Moritz RL, Sun Z, Deutsch EW, Gotthardt M, Mudge JM, Prensner JR, Willnow TE, Mertins P, Van Heesch S, Hubner N. Evolutionary origins and interactomes of human, young microproteins and small peptides translated from short open reading frames. Mol Cell 2023;83:994-1011.e18. doi: 10.1016/j.molcel.2023.01.023.
Litviňuková M#, Talavera-López C#, Maatz H, Reichart D#, Worth CL#, Lindberg EL, Kanda M, Polanski K, Heinig M, Lee M, Nadelmann ER, Roberts K, Tuck L, Fasouli ES, DeLaughter DM, McDonough B, Wakimoto H, Gorham JM, Samari S, Mahbubani KT, Saeb-Parsy K, Patone G, Boyle JJ, Zhang H, Zhang H, Viveiros A, Oudit GY, Bayraktar OA, Seidman JG, Seidman CE, Noseda M, Hubner N, Teichmann SA. Cells of the adult human heart. Nature 2020;588:466–72.doi: 10.1038/s41586-020-2797-4.
Reichart D#, Lindberg EL#, Maatz H#, Miranda AMA, Viveiros A, Shvetsov N, Gärtner A, Nadelmann ER, Lee M, Kanemaru K, Ruiz-Orera J, Strohmenger V, DeLaughter DM, Patone G, Zhang H, Woehler A, Lippert C, Kim Y, Adami E, Gorham JM, Barnett SN, Brown K, Buchan RJ, Chowdhury RA, Constantinou C, Cranley J, Felkin LE, Fox H, Ghauri A, Gummert J, Kanda M, Li R, Mach L, McDonough B, Samari S, Shahriaran F, Yapp C, Stanasiuk C, Theotokis PI, Theis FJ, Bogaerdt A van den, Wakimoto H, Ware JS, Worth CL, Barton PJR, Lee YA, Teichmann SA, Milting H#, Noseda M#, Oudit GY#, Heinig M#, Seidman JG#, Hubner N#, Seidman CE#. Pathogenic variants damage cell composition and single-cell transcription in cardiomyopathies. Science 2022;377:eabo1984–eabo1984. doi: 10.1126/science.abo1984.
Poch T, Krause J, Casar C, Liwinski T, Glau L, Kaufmann M, Ahrenstorf AE, Hess LU, Ziegler AE, Martrus G, Lunemann S, Sebode M, Li J, Schwinge D, Krebs CF, Franke A, Friese MA, Oldhafer KJ, Fischer L, Altfeld M, Lohse AW, Huber S, Tolosa E, Gagliani N, Schramm C. Single-cell atlas of hepatic T cells reveals expansion of liver-resident naive-like CD4+ T cells in primary sclerosing cholangitis. J Hepatol 2021;75(2):414–23. doi: 10.1016/j.jhep.2021.03.016.
Stein S, Henze L, Poch T, Carambia A, Krech T, Preti M, Schuran FA, Reich M, Keitel V, Fiorotto R, Strazzabosco M, Fischer L, Li J, Müller LM, Wagner J, Gagliani N, Herkel J, Schwinge D, Schramm C. IL-17A/F enable cholangiocytes to restrict T cell-driven experimental cholangitis by upregulating PD-L1 expression. J Hepatol 2021;74:919–30. doi: 10.1016/j.jhep.2020.10.035.
Zecher BF, Ellinghaus D, Schloer S, Niehrs A, Padoan B, Baumdick ME, Yuki Y, Martin MP, Glow D, Schröder-Schwarz J, Niersch J, Brias S, Müller LM, Habermann R, Kretschmer P, Früh T, Dänekas J, Wehmeyer MH, Poch T, Sebode M, (IPSCSG) IPSG, Ellinghaus E, Degenhardt F, Körner C, Hoelzemer A, Fehse B, Oldhafer KJ, Schumacher U, Sauter G, Carrington M, Franke A, Bunders MJ, Schramm C, Altfeld M. HLA-DPA1*02:01~B1*01:01 is a risk haplotype for primary sclerosing cholangitis mediating activation of NKp44+ NK cells. Gut 2024;73:325–37. doi: 10.1136/gutjnl-2023-329524.
Mudge JM#, Ruiz-Orera J#, Prensner JR#, Brunet MA, Calvet F, Jungreis I, Gonzalez JM, Magrane M, Martinez TF, Schulz JF, Yang YT, Albà MM, Aspden JL, Baranov PV, Bazzini AA, Bruford E, Martin MJ, Calviello L, Carvunis AR, Chen J, Couso JP, Deutsch EW, Flicek P, Frankish A, Gerstein M, Hubner N, Ingolia NT, Kellis M, Menschaert G, Moritz RL, Ohler U, Roucou X, Saghatelian A, Weissman JS, Van Heesch S#. Standardised annotation of translated open reading frames. Nat Biotechnol 2022;40:994–9. doi: 10.1038/s41587-022-01369-0.
# equally contributing authors