
_st_plus_slider_1680_600.png)
Metagenomics in emerging viral infections – integrating non-host and host transcriptional responses
Summary
Emerging viral infections remain a significant threat to public health, and their early detection is essential for effective containment and treatment. Traditional diagnostic methods, although advanced, often fall short in detecting novel or highly diverse pathogens - especially during the early stages of infection or when the pathogen is not present in easily accessible samples. Metagenomic next-generation sequencing (mNGS), particularly RNA-Seq, offers a promising solution by enabling unbiased detection of pathogen transcripts and simultaneous analysis of the host’s gene expression. This dual capability is particularly useful in acute infections where high levels of pathogen replication occur, as well as for identifying host response patterns that may point to infection even in the absence of detectable pathogen genetic material.
This project will build on existing expertise in viral genomics and metagenomics to improve pathogen detection and distinguish infections from other inflammatory conditions. A key goal is to combine pathogen data with host response signatures to create a more accurate diagnostic fingerprint - helping to differentiate active infections from incidental colonization. The project will also evaluate the use of short host gene expression signatures to distinguish viral from bacterial infections in both systemic and local contexts, such as respiratory or cerebrospinal fluid samples.
Furthermore, it aims to develop host-specific response patterns for particular viral families, exemplified by the Arenaviridae. All findings will be integrated into the existing DAMIAN pathogen pipeline to make these advanced diagnostic tools accessible to the broader research and clinical community. Ultimately, this work aims to enable earlier and more accurate detection of known and unknown pathogens, including the hypothetical “Pathogen X,” by leveraging both host and pathogen data.
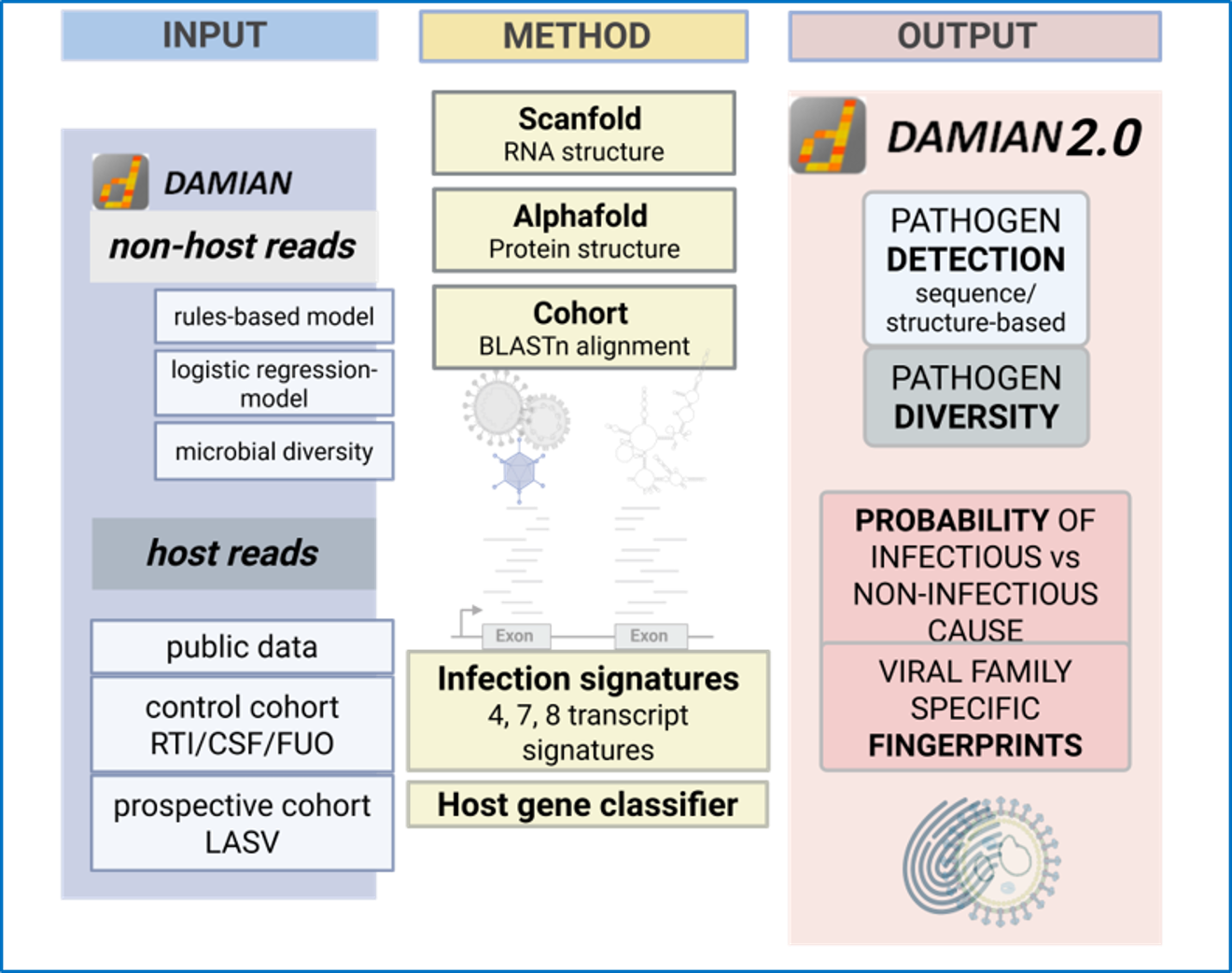
Our Work Packages (WP):
WP01: Identification of viruses using sequence and structural information – DAMIAN 2.0
We investigate how to improve virus detection by integrating RNA secondary structure prediction (ScanFold) and protein structure modeling (AlphaFold/ESMFold) into the DAMIAN pipeline to identify viral families beyond primary sequence similarity.
WP02: Identification of a shared host response signature in different diagnostic specimens
We investigate whether small host gene expression signatures can distinguish between viral and bacterial infections across different sample types such as CSF, BAL, and blood.
WP03: Identification and validation of LASV- and filovirus-specific host response signatures
We investigate whether host transcriptome data can be used to define and validate virus family-specific immune response patterns, using LASV as a model.
With these work packages, we aim to develop a comprehensive strategy for early and sequence-independent detection of viral infections and pathogen-specific host responses.
Team


Prof. Dr. Nicole Fischer
Principal Investigator
E-mail address:
Phone: +49 (0) 40 7410 - 55171
Institute for Molecular Virology and Tumorvirology (MVTV)
The Institute for Molecular Virology and Tumorvirology led by Prof. Dr. Fischer is based at the University Medical Center Hamburg-Eppendorf.
Project related publications
Heyer A, Gunther T, Robitaille A, …, Addo MM, …, Schulze zur Wiesch J, Fischer N#, Grundhoff A#. Remdesivir-induced emergence of SARS-CoV-2 variants in patients with prolonged infection. Cell Rep Med. 2022;3(9):100735. doi: 10.1016/j.xcrm.2022.100735
Pfefferle S, Gunther T, Kobbe R, …, Aepfelbacher M#, Knobloch J#, Lütgehetmann M#, Fischer N#. SARS Coronavirus-2 variant tracing within the first Coronavirus Disease 19 clusters in northern Germany. Clin Microbiol Infect. 2021;27(1):130.e5–130.e8. doi: 10.1016/j.cmi.2020.09.034
Gunther T, Czech-Sioli M, Indenbirken D, …, Fischer N#, Grundhoff A#, Brinkmann M#. SARS-CoV-2 outbreak investigation in a German meat processing plant. EMBO Mol Med. 2020;12(12):e13296. doi: 10.15252/emmm.202013296
Czech-Sioli M, Gunther T, Therre M, …, Becker J, Grundhoff A#, Fischer N#. High-resolution analysis of Merkel Cell Polyomavirus in Merkel Cell Carcinoma reveals distinct integration patterns and suggests NHEJ and MMBIR as underlying mechanisms. PLoS Pathog. 2020;16(8):e1008562. doi: 10.1371/journal.ppat.1008562
Siebels S, Czech-Sioli M, Spohn M, …, Indenbirken D, Grundhoff A, Fischer N. Merkel Cell Polyomavirus DNA Replication Induces Senescence in Human Dermal Fibroblasts in a Kap1/Trim28-Dependent Manner. mBio. 2020;11(2):e00142-20. doi: 10.1128/mBio.00142-20
Alawi M, Burkhardt L, Indenbirken D, …, Lütgehetmann M, Aepfelbacher M, Fischer N#, Grundhoff A#. DAMIAN: an open source bioinformatics tool for fast, systematic and cohort based analysis of microorganisms in diagnostic samples. Sci Rep. 2019;9(1):16841. doi: 10.1038/s41598-019-52881-4
Postel A, Meyer D, Cagatay GN, …, Fischer N, …, Sun H, Wendt M, Becher P. High Abundance and Genetic Variability of Atypical Porcine Pestivirus in Pigs from Europe and Asia. Emerg Infect Dis. 2017;23(12):2104–2107. doi: 10.3201/eid2312.170951
Baechlein C#, Fischer N#, Grundhoff A, …, Beineke A, Rehage J, Becher P. Identification of a Novel Hepacivirus in Domestic Cattle from Germany. J Virol. 2015;89(14):7007–7015. doi: 10.1128/JVI.00534-15
Fischer N#, Indenbirken D, Meyer T, …, Aepfelbacher M, Alawi M, Grundhoff A#. Evaluation of Unbiased Next-Generation Sequencing of RNA (RNA-seq) as a Diagnostic Method in Influenza Virus-Positive Respiratory Samples. J Clin Microbiol. 2015;53(7):2238–2250. doi: 10.1128/JCM.02495-14
Fischer N#, Rohde H, Indenbirken D, …, Gunther T, Aepfelbacher A, Grundhoff A#. Rapid metagenomic diagnostics for suspected outbreak of severe pneumonia. Emerg Infect Dis. 2014;20(6):1072–1075. doi: 10.3201/eid2006.131526
#equally contributing authors