
SP01:
Discovering liver pathomechanisms via computational and statistical analysis support and training
Project leaders:
Prof. Dr. rer. nat. Stefan Bonn
Dr. rer. nat. Olga Zolotareva
Prof. Dr. rer. nat. Jan Baumbach



Summary
The main aim of this CRC is to mechanistically understand immune regulation in the liver in order to develop novel treatment approaches. We hypothesise that the modelling of liver phenotypes across high-throughput data, diseases and data types will help us to understand how the immune system, genetic factors and clinical factors predict, explain and stratify liver diseases. SP01 will provide the necessary bioinformatic and statistical expertise to support CRC researchers with best-of-breed analysis routines throughout the data lifecycle.
Over the past decades, the rapid growth of high-throughput technologies has yielded a large volume of biomedical data that enables unprecedented insights into biological processes. At the same time, this development has shifted the bottleneck of research from data generation to data integration and interpretation. In this CRC initiative, a variety of high-throughput omics data in addition to clinical, paraclinical and biosample-associated data will be generated and integrated. Managing this cross-sectional, cross-institutional and multidisciplinary endeavour poses a challenge that is addressed in project INF. Project SP01 will utilise the data provided by the INF project to i) provide state-of-the-art data analysis support, ii) consult and train CRC researchers in computational data analysis and statistics and iii) develop novel AI-based methods for high-throughput, high-dimensional data analysis, feature extraction and patient stratification.
The main aim of algorithmic developments will be to delineate the molecular mechanisms and potential treatment targets underlying hepatic autoimmune and immune-mediated diseases, cancer and infections. Moreover, SP01 will support the Academy for Translational Liver Immunology (iRTG) to enable knowledge transfer of bioinformatics skills to the next generation of biomedical and clinical scientists.
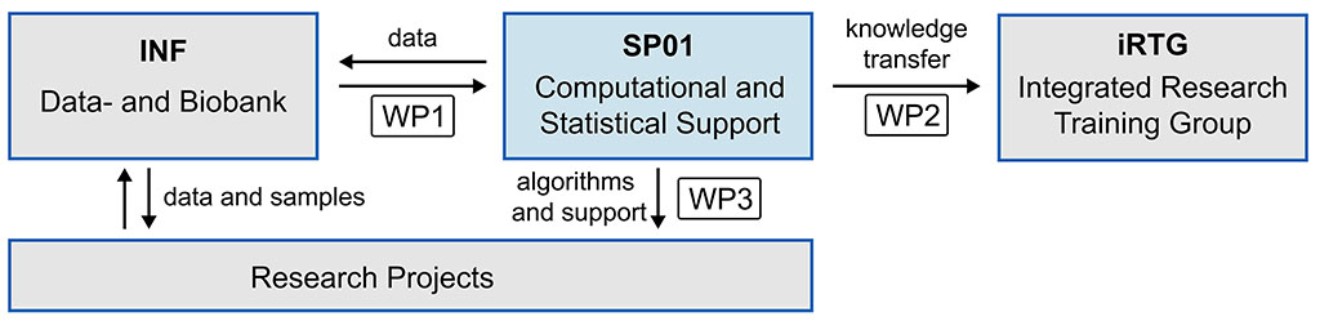
Project plan
SP01 will support the i) computational data analysis across projects and data types, ii) consult and teach in bioinformatics and statistics and iii) develop state-of-the-art algorithms where required (see Fig. below). These efforts will enable CRC researchers to decipher the molecular mechanisms underlying hepatic disease from complex data, identify disease mechanisms, stratify patients based on molecular signatures and enable personalised treatment regimes. Efficient data access of SP01 will be provided by Ückert & Sebode (INF), who lead the data- and biobank project. Knowledge transfer will be guaranteed by closely collaborating with the iRTG led by Schwinge & Lohse.
Project related publications
List M, Alcaraz N, Dissing-Hansen M, Ditzel HJ, Mollenhauer J, Baumbach J. KeyPathwayMinerWeb: online multi-omics network enrichment. Nucleic Acids Re 2016;44(W1):W98-W104. doi: 10.1093/nar/gkw373. Epub 2016 May 5. Open access.
Larsen SJ, Schmidt HHHW, Baumbach J. De novo and supervised endophenotyping using network-guided ensemble learning. Systems Medicine 2020 3:1,8–21. Open access.
Zolotareva O, Khakabimamaghani S, Isaeva OI, Chervontseva Z, Savchik A, Ester M. Identification of differentially expressed gene modules in heterogeneous diseases. Bioinformatics 2021;37:1691-1698. doi: 10.1093/bioinformatics/btaa1038.
Zolotareva O, Isaeva OI, Hartung M, Hartung M, Maier A, Delgado-Chaves FM, Kaufmann K, Savchik A, Chervontseva Z, Probul N, Abisheva A, Zotova E, Tsoy O, Blumenthal DB, Ester M, Baumbach J. DESMOND 2.0: Identification of differentially expressed biclusters for unsupervised patient stratification. ScienceOpen 2022. doi:10.14293/S2199-1006.1.SOR-.PPPSLHRB.v1.
Sadegh S#, Skelton J#, Anastasi E, Bernett J, Blumenthal DB, Galindez G, Salgado-Albarrán M, Lazareva O, Flanagan K, Cockell S, Nogales C, Casas AI, Schmidt HHHW, Baumbach J#, Wipat A#, Kacprowski T#. Network medicine for disease module identification and drug repurposing with the NeDRex platform. Nat Commun 2021;12:6848. doi: 10.1038/s41467-021-27138-2. Open access.
Maier A#, Hartung M, Abovsky M, Adamowicz K, Bader GD, Baier S, Blumenthal DB, Chen J, Elkjaer ML, Garcia-Hernandez C, Helmy M, Hoffmann M, Jurisica I, Kotlyar M, Lazareva O, Levi H, List M, Lobentanzer S, Loscalzo J, Malod-Dognin N, Manz Q, Matschinske J, Mee M, Oubounyt M, Pastrello C, Pico AR, Pillich RT, Poschenrieder JP, Pratt D, Pržulj N, Sadegh S, Saez-Rodriguez J, Sarkar S, Shaked G, Shamir R, Trummer N, Turhan U, Wang RS, Zolotareva O, Baumbach J. Drugst.One — a plug-and-play solution for online systems medicine and network-based drug repurposing, Nucleic Acids Research, 2024;Vol. 52; Issue W1:W481 W488, https://doi.org/10.1093/nar/gkae388.
Ragab H#, Westhaeusser F#, Ernst A, Yamamura J, Fuhlert P, Zimmermann M, Sauerbeck J, Shenas F, Özden C, Weidmann A, Adam G, Bonn S#, Schramm C#. DeePSC: A Deep Learning Model for Automated Diagnosis of Primary Sclerosing Cholangitis on 2D MR Cholangiopancreatography. Radiol Artif Intell 2023;5:e220160. doi:10.1148/ryai.220160.
Kylies D, Zimmermann M, Haas F, Schwerk M, Kuehl M, Brehler M, Czogalla J, Hernandez LC, Konczalla L, Okabayashi Y, Menzel J, Edenhofer I, Mezher S, Aypek H, Dumoulin B, Wu H, Hofmann S, Kretz O, Wanner N, Tomas NM, Krasemann S, Glatzel M, Kuppe C, Kramann R, Banjanin B, Schneider RK, Urbschat C, Arck P, Gagliani N, van Zandvoort M, Wiech T, Grahammer F, Sáez PJ, Wong MN, Bonn S, Huber TB, Puelles VG. Expansion-enhanced super-resolution radial fluctuations enable nanoscale molecular profiling of pathology specimens. Nat Nanotechnol 2023;18:336-342. doi: 10.1038/s41565-023-01328-z. Epub 2023 Apr 10. Open access.
Hausmann F#, Ergen C#, Khatri R, Marouf M, Hänzelmann S, Gagliani N, Huber S, Machart P, Bonn S. DiSCERN: deep single-cell expression reconstruction for improved cell clustering and cell subtype and state detection. Genome Biology 2023;24:212. doi:10.1186/s13059-023-03049-x. Open access.
Khatri R, Machart P, Bonn S. Semi-supervised consistency regularization for accurate cell type fraction and gene expression estimation. PREPRINT available at Research Square 2023. doi.org/10.21203/rs.3.rs-2573385/v1.
# equally contributing authors