 Research at the UKE?
Research at the UKE?
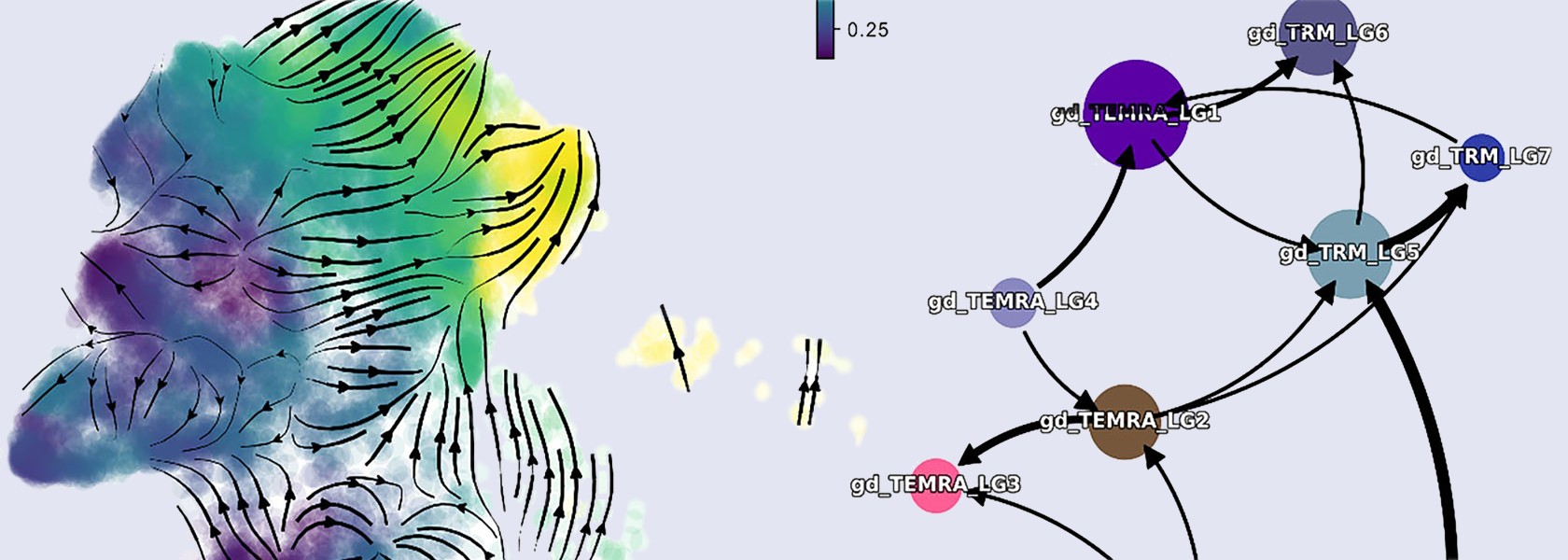
Systems Immunology
Our goal is to understand the immunological opportunities and challenges arising from the enormous diversity of adaptive T cell receptor combinations. Therefore, we monitor the dynamics of T cell receptor repertoires during immune responses to pathogens and in the course of autoimmune diseases.
In particular, we are interested in the role of γδ T cells at the interface between innate and adaptive immunity.

“Unraveling the mysteries of γδ T cells.”
Prof. Dr. Immo Prinz
Project details and goals
T cells develop in the thymus, where each of them rearranges a different αβ or γδ T cell receptor. The theoretical diversity of these T cell receptors is more than a quadrillion, which allows for the potential recognition of almost infinite numbers of antigens by different T cell receptors. We hypothesize that the individual T cell receptor sequences determine the functional differentiation of αβ or γδ T cells.
To understand the correlation between T cell receptor sequences and immune responses, our specific goals are:
- to track individual T cell clones that respond to viral infections by single-cell sequencing in longitudinal cohorts;
- to identify the antigens recognized by responding αβ or γδ T cell receptors using experimental and in silico systems;
- to study the clonal distribution of T cells and their interaction with target cells in tissues.
In summary, we aim to understand how individual T cell receptors instruct αβ and γδ T cells to mount a beneficial response to infection and cancer, or to eventually drive adverse autoimmune reactions.
In the long run, the “good” T cell receptors could be used for immunotherapy and the “bad” T cell receptors targeted to dampen autoimmunity.
Current projects
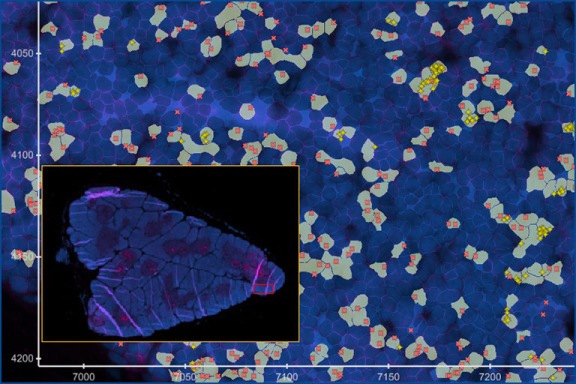
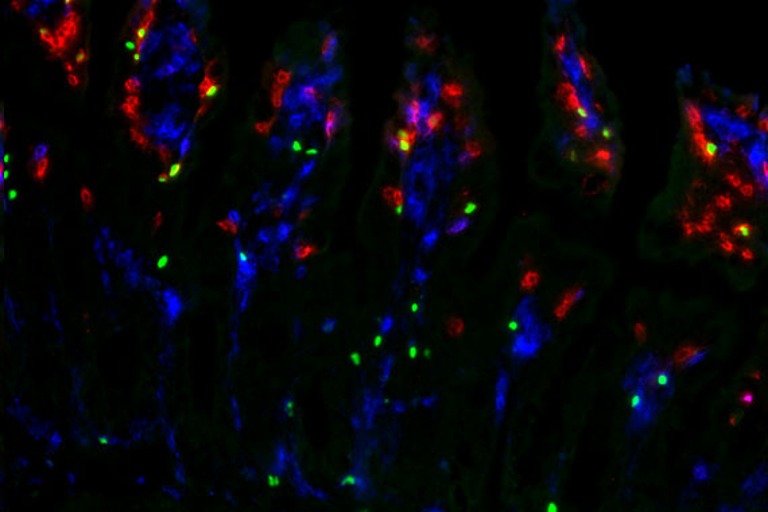
Clonal distribution of T cells and their interaction with target cells in tissues
Description: The 10X Xenium in situ platform enables subcellular resolution for up to 500 genes within a single panel. We designed a panel to detect all variable “V” genes of αβ and γδ T cell receptors and identify T cell subsets. By analyzing the spatial distribution of TRAV and TRBV gene combinations, we can identify microrepertoires in tissues and lymphatic organs.
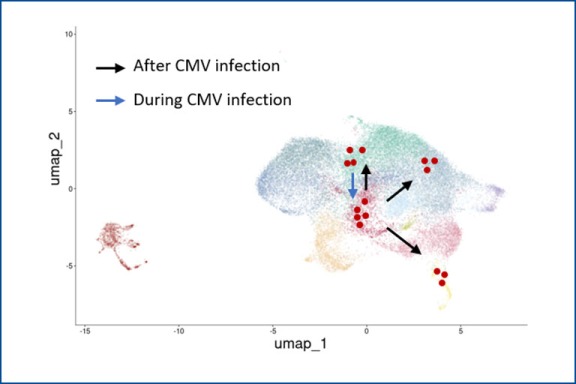
Tracking individual γδ and αβ T cell clones during CMV reactivation
Description: In this project, funded by the EU consortium HORUS, we analyze the immune response of γδ and αβ T cells to cytomegalovirus (CMV) infection and reactivation. We use cell sorting, flow cytometry, single-cell RNA sequencing, and single-cell TCR sequencing to track clonal expansion and phenotypic changes and in γδ and αβ T cells at different stages: before, during, and after CMV reactivation. Individual T cell clones are identified by their unique CDR3 sequences.
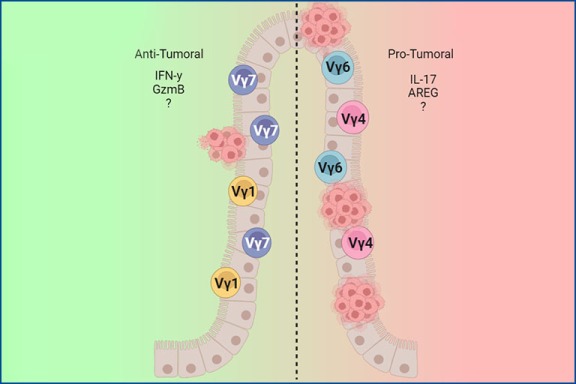
Contribution of γδ T cells to the development of colitis-associated colorectal cancer
The colon harbors one of the largest immune cell compartments in the entire organism. In its mucosa, γδ T cells can be found either as intraepithelial intestinal T cells (IEL), located within the thin epithelial layer, or as lamina propria resident cells (LPL), primarily exhibiting an effector phenotype characterized mainly by IL-17 production. Chronic inflammation of the intestine can lead to a remodeling of this immune cell compartment, thus compromising the maintenance and homeostasis of the organ. Ultimately, chronic inflammation can lead to the development of colorectal cancer (CRC). Our goal is to outline the extent to which γδ T cells are involved in the development of colitis-associated CRC. More specifically, we are interested in understanding whether γδ T cell function during the course of this disease is associated with specific cell clones and whether these clones accumulate in the tumor. Finally, we aim to elucidate which changes during tumorigenesis potentially lead to the recognition of malignant cells by γδ T cells, and what kind of cytotoxic machinery is involved in killing transformed cells.
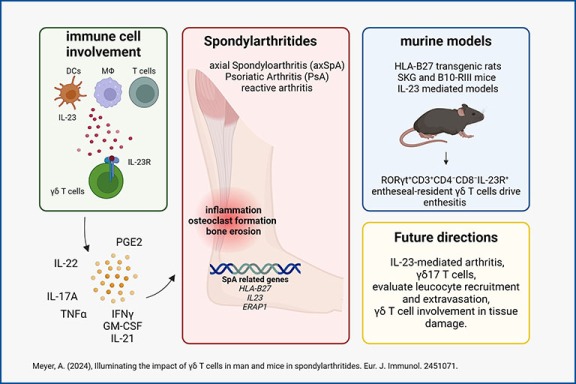
Deciphering γδ T cells in spondyloarthropathies
γδ T cells in spondyloarthropathies (SpA) have a distinct phenotype, expressing the IL-23 receptor (IL-23R) and the transcription factor RORγt. IL-23/IL-23R signaling is essential for their activation in SpA. However, the specific cells and factors involved in IL-23-mediated immune reactions remain unclear. Despite effective drugs for psoriatic arthritis that target the IL-23 pathway, IL-23R inhibition is ineffective in SpA. Murine models provide valuable insights into the progression of SpA. We hypothesize that γδ T cells are pivotal in SpA pathogenesis by driving inflammation, tissue damage, and modulating leukocyte recruitment.
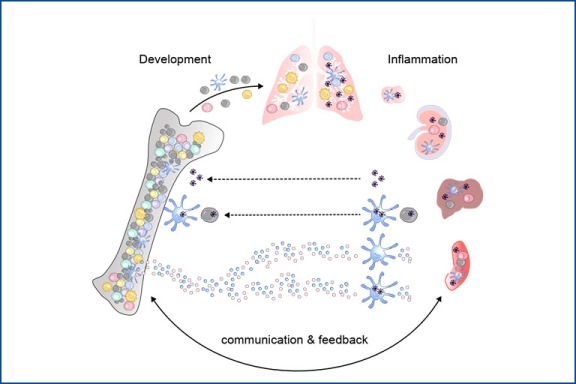
Development of functionally diverse myeloid cells in steady state vs. disease
This project focuses on the ontogeny of mononuclear phagocytes – what drives the differentiation of true cell subsets versus functional cell states in the bone marrow versus tissues. Since development and functional imprinting are closely linked processes, we are investigating how perturbations by viral infection, autoimmunity or cancer affect these decisions. We want to understand how the tissue niche influences the functional decisions of mononuclear phagocytes and their progenitors, their cell-cell interactions in distinct tissues and disease contexts, how these functions are controlled and via which mechanisms inflammatory signals from the tissue reach the bone marrow altering the progenitor pool. Given the unmatched efficiency of dendritic cells in presenting antigens to T cells and to induce sustainable immune responses, these cells are intriguing targets for improved vaccines and immunotherapies. Thus, if we want to ‘exploit’ the properties of these cells and their progenitors, we first need to fully understand their biology. To address our research questions, we combine immunological and genetic methods, single-cell genomics, spatial transcriptomics, and high-parameter flow cytometry with bioinformatics approaches.
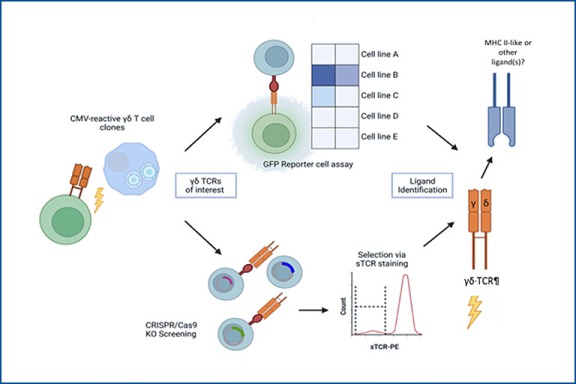
Identification of γδ T cell receptor ligands
In contrast to αβ T cells, γδ T cells are not usually restricted to peptide:MHC recognition. A concept for antigen recognition of γδ TCR is missing and only very few and diverse ligands are known. In this project, we aim to find the cognate ligand(s) of γδ TCRs that expanded clonally after, for example, CMV infection or reactivation. We use in vitro approaches, such as a JE6.1-NFKB-GFP Jurkat reporter cell line and soluble domains of the γδ TCRs (sTCR) to screen for reactivity against potential target cell lines. Combined with sorting and cloning of confirmed target cells that show loss of binding after a CRISPR/Cas9 genome-wide knockout screen, we aim to elucidate candidate genes encoding for unknown ligand(s) of γδ T cells. This study could further shed light on the complex ligand recognition patterns of γδ T cells and exploit these for translational research.
Publications
Deseke M, Rampoldi F, Sandrock I, Borst E, Böning H, Ssebyatika GL, Jürgens C, Plückebaum N, Beck M, Hassan A, Tan L, Demera A, Janssen A, Steinberger P, Koenecke C, Viejo-Borbolla A, Messerle M, Krey T, Prinz I. J Exp Med. 2022. doi: 10.1084/jem.20212525.
Abstract
The innate and adaptive roles of γδ T cells and their clonal γδ T cell receptors (TCRs) in immune responses are still unclear. Recent studies of γδ TCR repertoire dynamics showed massive expansion of individual Vδ1+ γδ T cell clones during viral infection. To judge whether such expansion is random or actually represents TCR-dependent adaptive immune responses, information about their cognate TCR ligands is required. Here, we used CRISPR/Cas9-mediated screening to identify HLA-DRA, RFXAP, RFX5, and CIITA as required for target cell recognition of a CMV-induced Vγ3Vδ1+ TCR, and further characterization revealed a direct interaction of this Vδ1+ TCR with the MHC II complex HLA-DR. Since MHC II is strongly upregulated by interferon-γ, these results suggest an inflammation-induced MHC-dependent immune response of γδ T cells.
Tan L, Fichtner AS, Bruni E, Odak I, Sandrock I, Bubke A, Borchers A, Schultze-Florey C, Koenecke C, Förster R, Jarek M, von Kaisenberg C, Schulz A, Chu X, Zhang B, Li Y, Panzer U, Krebs CF, Ravens S*, Prinz I*. Sci Immunol. 2021. doi: 10.1126/sciimmunol.abf0125, *Equal contribution.
Abstract
Accumulating evidence suggests that the mouse embryonic thymus produces distinct waves of innate effector γδ T cells. However, it is unclear whether this process occurs similarly in humans and whether it comprises a dedicated subset of innate-like type 3 effector γδ T cells. Here, we present a protocol for high-throughput sequencing of TRG and TRD pairs that comprise the clonal γδTCR. In combination with single-cell RNA sequencing, multiparameter flow cytometry, and TCR sequencing, we reveal a high heterogeneity of γδ T cells sorted from neonatal and adult blood that correlated with TCR usage. Immature γδ T cell clusters displayed mixed and diverse TCRs, but effector cell types segregated according to the expression of either highly expanded individual Vδ1+ TCRs or moderately expanded semi-invariant Vγ9Vδ2+ TCRs. The Vγ9Vδ2+ T cells shared expression of genes that mark innate-like T cells, including ZBTB16 (encoding PLZF), KLRB1, and KLRC1, but consisted of distinct clusters with unrelated Vγ9Vδ2+ TCR clones characterized either by TBX21, FCGR3A, and cytotoxicity-associated gene expression (type 1) or by CCR6, RORC, IL23R, and DPP4 expression (type 3). Effector γδ T cells with type 1 and type 3 innate T cell signatures were detected in a public dataset of early embryonic thymus organogenesis. Together, this study suggests that functionally distinct waves of human innate-like effector γδ T cells with semi-invariant Vγ9Vδ2+TCR develop in the early fetal thymus and persist into adulthood.
Ravens S, Fichtner AS, Willers M, Torkornoo D, Pirr S, Schöning J, Deseke M, Sandrock I, Bubke A, Wilharm A, Dodoo D, Egyir B, Flanagan KL, Steinbrück L, Dickinson P, Ghazal P, Adu B, Viemann D*, Prinz I*. Proc Natl Acad Sci U S A. 2020. doi: 10.1073/pnas.1922588117, *Equal contribution.
Abstract
Starting at birth, the immune system of newborns and children encounters and is influenced by environmental challenges. It is still not completely understood how γδ T cells emerge and adapt during early life. Studying the composition of T cell receptors (TCRs) using next-generation sequencing (NGS) in neonates, infants, and children can provide valuable insights into the adaptation of T cell subsets. To investigate how neonatal γδ T cell repertoires are shaped by microbial exposure after birth, we monitored the γ-chain (TRG) and δ-chain (TRD) repertoires of peripheral blood T cells in newborns, infants, and young children from Europe and sub-Saharan Africa. We identified a set of TRG and TRD sequences that were shared by all children from Europe and Africa. These were primarily public clones, characterized by simple rearrangements of Vγ9 and Vδ2 chains with low junctional diversity and usage of non-TRDJ1 gene segments, reminiscent of early ontogenetic subsets of γδ T cells. Further profiling revealed that these innate, public Vγ9Vδ2+ T cells underwent an immediate TCR-driven polyclonal proliferation within the first 4 wk of life. In contrast, γδ T cells using Vδ1+ and Vδ3+ TRD rearrangements did not significantly expand after birth. However, different environmental cues may lead to the observed increase of Vδ1+ and Vδ3+ TRD sequences in the majority of African children. In summary, we show how dynamic γδ TCR repertoires develop directly after birth and present important differences among γδ T cell subsets.
Sandrock I, Reinhardt A, Ravens S, Binz C, Wilharm A, Martins J, Oberdörfer L, Tan L, Lienenklaus S, Zhang B, Naumann R, Zhuang Y, Krueger A, Förster R, Prinz I. J Exp Med. 2018. doi: 10.1084/jem.20181439.
Abstract
γδ T cells are highly conserved in jawed vertebrates, suggesting an essential role in the immune system. However, γδ T cell-deficient Tcrd−/− mice display surprisingly mild phenotypes. We hypothesized that the lack of γδ T cells in constitutive Tcrd−/− mice is functionally compensated by other lymphocytes taking over genuine γδ T cell functions. To test this, we generated a knock-in model for diphtheria toxin–mediated conditional γδ T cell depletion. In contrast to IFN-γ–producing γδ T cells, IL-17–producing γδ T cells (Tγδ17 cells) recovered inefficiently after depletion, and their niches were filled by expanding Th17 cells and ILC3s. Complementary genetic fate mapping further demonstrated that Tγδ17 cells are long-lived and persisting lymphocytes. Investigating the function of γδ T cells, conditional depletion but not constitutive deficiency protected from imiquimod-induced psoriasis. Together, we clarify that fetal thymus-derived Tγδ17 cells are nonredundant local effector cells in IL-17–driven skin pathology.
Ravens S, Schultze-Florey C, Raha S, Sandrock I, Drenker M, Oberdörfer L, Reinhardt A, Ravens I, Beck M, Geffers R, von Kaisenberg C, Heuser M, Thol F, Ganser A, Förster R, Koenecke C*, Prinz I*. Nat Immunol. 2017. doi: 10.1038/ni.3686, *Equal contribution.
Abstract
To investigate how the human γδ T cell pool is shaped during ontogeny and how it is regenerated after transplantation of hematopoietic stem cells (HSCs), we applied an RNA-based next-generation sequencing approach to monitor the dynamics of the repertoires of γδ T cell antigen receptors (TCRs) before and after transplantation in a prospective cohort study. We found that repertoires of rearranged genes encoding γδ TCRs (TRG and TRD) in the peripheral blood of healthy adults were stable over time. Although a large fraction of human TRG repertoires consisted of public sequences, the TRD repertoires were private. In patients undergoing HSC transplantation, γδ T cells were quickly reconstituted; however, they had profoundly altered TCR repertoires. Notably, the clonal proliferation of individual virus-reactive γδ TCR sequences in patients with reactivation of cytomegalovirus revealed strong evidence for adaptive anti-viral γδ T cell immune responses.
Team


Prof. Dr. Immo Prinz
Head
E-mail address:




Dr. rer. nat. Regine Dress
Junior Research Group Leader
The Myeloid Systems Immunology Lab focuses on the ontogeny of mononuclear phagocytes – what drives cell fate decisions in the bone marrow vs. tissues. As development and functional imprinting are closely tied together processes, we are investigating how perturbation, e.g. by viral infections and the cellular environment impacts these decisions. To address these questions, we combine single-cell genomics, spatial transcriptomics, and high-parameter flow cytometry with machine learning approaches.
Contact:




